
Therapeutic Area User Guide for HIV
Version 1.0 (Provisional)
Notes to Readers
- This is the provisional version 1.0 of the Therapeutic Area User Guide for HIV.
- This document is based on SDTM v1.7, SDTMIG v3.3 CDASH Model v1.0, CDASHIG v2.0, SDTMIG-AP v1.0, SDTMIG-MD v1.1, SDTMIG-PGx v1.0, ADaMIG v1.1 and ADaM v2.1.
Revision History
| Date | Version |
|---|---|
| 2019-01-18 | 1.0 Provisional |
© 2019 Clinical Data Interchange Standards Consortium, Inc. All rights reserved.
Contents
- 1 Introduction
- 2 Overview of HIV
- 3 Subject and Disease Characteristics
- 4 Baseline Medical History and Physical Exams of Special Interest
- 5 Mother-Infant Pairs in HIV Studies
- 6 Interventions for the Prevention and Treatment of HIV
- 6.1 HIV Treatment with Oral Antiretroviral Therapy (ART)
- 6.2 Pre-exposure Prophylaxis (PrEP)
- 6.2.1 Vaginal Ring
- 6.3 Vaccines
- 6.3.1 Reactogenicity
- 6.3.2 Immune Response
- 6.3.3 Vaccine-induced Seroreactivity
- 7 Disease Assessments
- 8 Analysis Data
- Appendices
- Appendix A: HIV Standards Team
- Appendix B: Glossary and Abbreviations
- Appendix C: Non-Standard Variables
- Appendix D: References
- Appendix E: Representations and Warranties, Limitations of Liability, and Disclaimers
1 Introduction
This Therapeutic Area Data Standards User Guide for HIV (TAUG-HIV) was developed under the Coalition for Accelerating Standards and Therapies (CFAST) initiative.
The purpose of the TAUG-HIV is to describe how to use Clinical Data Interchange Standards Consortium (CDISC) standards to represent data pertaining to HIV or data commonly collected in HIV trials. These include studies of both HIV treatment and HIV prevention, as well as studies that may include HIV-infected individuals.
1.1 How to Read this Document
- First, read the Study Data Tabulation Model (SDTM) Version 1.7, SDTM Implementation Guide (SDTMIG) Version 3.3, Clinical Data Acquisition Standards Harmonization (CDASH) Model Version 1.0, CDASH Implementation Guide (CDASHIG) Version 2.0, Analysis Data Model (ADaM) Version 2.1, and ADaM Implementation Guide (ADaMIG) Version 1.1. These standards are available at http://www.cdisc.org/.
- As needed, visit Section 1.3, CDASH Metadata and Annotated CRFs, of this guide for an explanation of how to read the annotated CRF.
- Read the SDTMIG for Associated Persons (SDTMIG-AP), SDTMIG for Pharmacogenomics/Genetics (SDTMIG-PGx), and SDTMIG for Medical Devices (SDTMIG-MD) to gain some familiarity with representing non-study subjects, genetics, and medical devices in the SDTM. These standards are available from: http://www.cdisc.org/.
- Read the Introduction to Therapeutic Area Standards (http://wiki.cdisc.org/x/SSy8AQ) and/or take CDISC's free training module, TA001 - Overview of Therapeutic Area User Guides (https://www.cdisc.org/education), to understand what to expect from a therapeutic area user guide.
- Read this guide all the way through (without skipping any sections) at least once.
- Finally, revisit any sections of particular interest.
Draft standards of interest to this document are listed at: Draft Standards of Interest to TAUG-HIV ( https://wiki.cdisc.org/x/OJ2FAg ).
For information and specifications for the content of data sets that should be submitted as part of the sponsor or applicant's application for drugs intended to treat HIV, refer to the FDA's HIV Technical Specifications Guidance: https://www.fda.gov/downloads/ForIndustry/DataStandards/StudyDataStandards/UCM603323.pdf.
All general caveats for the standards given in the Introduction to Therapeutic Area Standards (http://wiki.cdisc.org/x/SSy8AQ) apply to this document.
Implementers should always verify terminology shown in examples against current, published CDISC Controlled Terminology, available at the National Cancer Institute website (https://www.cancer.gov/research/resources/terminology/cdisc).
1.2 Organization of this Document
This document is divided into the following sections:
- Section 1, Introduction, provides an overall introduction to the purpose and goals of the HIV project.
- Section 2, Overview of HIV, provides an overview of what HIV is and whom it affects.
- Section 3, Subject and Disease Characteristics, covers data that are usually collected once at the beginning of a study.
- Section 4, Baseline Medical History and Physical Exams of Special Interest, contains examples and discussion on baseline menstrual history and pelvic exam.
- Section 5, Mother-Infant Pairs in HIV Studies, covers the management of mother-infant relationships, with respect to the representation of data collected on either the mother or infant or both.
- Section 6, Interventions for the Prevention and Treatment of HIV, contains information about and examples of HIV interventions.
- Section 7, Disease Assessments, provides information on data that are typically collected multiple times during a study.
- Section 8, Analysis Data, includes key data analysis concepts for an HIV study.
- Appendices provide additional background material and describe other supplemental material relevant to HIV.
1.3 CDASH Metadata and Annotated CRFs
CDASH examples include both metadata tables and sample case report forms (CRFs). Each table of CDASH metadata corresponds to an example annotated CRF (aCRF), built directly from the metadata. The annotations show the variables associated with each field in the context of data collection (CDASH) and submission (SDTM). The color of the variable name denotes the applicable context. SDTMIG variable names are shown in RED. If the same variable name is used for both the SDTMIG variable and the CDASHIG variable, only the SDTMIG variable is shown. If the CDASHIG variable differs from the one defined in the SDTMIG, the CDASHIG variable is also shown in GRAY. Data collected but not submitted in SDTM-based datasets are denoted as. Not submitted
The following diagram illustrates how to interpret the annotations.
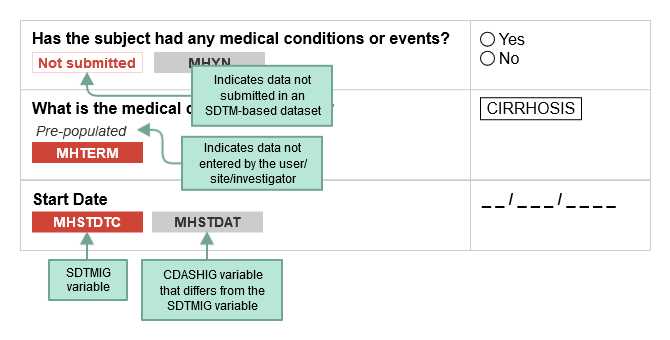
When viewing sample aCRFs, bear in mind that:
- Sample CRFs are provided to illustrate data collection instruments. These examples are not meant to imply that any particular layout is preferable over another.
- Example CRFs include annotations that refer to SDTM and CDASH data elements. Refer to these standards for more information about the use of these elements (https://www.cdisc.org/standards/foundational).
- Most example CRFs do not include header information such as subject identifier and visit, where applicable. Information in these headers may be populated in a variety of ways depending on the data management system.
- Sponsors are responsible for understanding and implementing CDISC Controlled Terminology (https://www.cancer.gov/research/resources/terminology/cdisc) where applicable.
- CDASH variable names for denormalized variables are examples. Sponsors may use other conventions for creating denormalized CDASH names.
- SDTM variable names separated by "/" indicate that either SDTM variable may be used when creating the SDTM datasets.
When viewing the CRF metadata, bear in mind that:
- Some CDASH metadata attributes are not included.
- Some non-CDASH metadata attributes necessary for rendering the CRF are included (eg, Hidden, Pre-specified Values, etc).
1.4 Known Issues
-
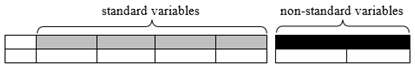
Non-standard variables: This document has adopted the practices outlined in the proposed SDTMIG Section 8.4.4, Alternative Representation of Non-Standard Variables (available in draft form at: http://wiki.cdisc.org/x/Ui68AQ). Accordingly, SDTM-based examples containing sample data requiring the use of a variable outside the standard set of variables included in the SDTM v1.7 are represented not with Supplemental Qualifier records, but with non-standard variables (NSVs) appended to the parent domain, followed by sample variable-level metadata for the NSVs.
In order to avoid confusion between standard variables and NSVs, NSVs have been rendered visually distinct, as shown below, with white text on black in the header row, and separated from the standard variables by a small space. Metadata for the NSVs, from the define.xml file that would accompany the submission, are tabulated below the example. Only those attributes or elements that assist the example are included. (For more information on variable-level metadata in general, see the Define-XML Specifications found at: https://www.cdisc.org/standards/data-exchange/define-xml ). A list of all NSVs used in this document, and the variable-level metadata that might become normative for the NSVs should they be promoted to standard variables, is given in Appendix C, Non-Standard Variables.
- Representation of prespecified findings: This guide shows an example in which prespecified findings from a pelvic exam are represented (see Section 4.2, Pelvic Examination). Within the CDISC community there is an ongoing discussion regarding the best way to represent prespecified findings. Historically, prespecified findings have been coordinated in the --TESTCD/--TEST variables (i.e., the prespecified target of interest is named in an indicator TEST, and the result is simply Y/N to indicate whether or not it was found). However, this can lead to a proliferation of controlled terms for these variables. Other options, including a new variable to represent the prespecified finding of interest (--RESTRG), are currently being explored. Previous therapeutic area user guides (TAUGs) have represented prespecified findings in the NSV --RESTRG. However, there has been a lack of consensus regarding this approach. Due to the lack of consensus, TAUGs will continue to coordinate prespecified findings in the --TESTCD/--TEST variables. This decision may change in the future.
-
Representation of subject characteristics collected more than once during a study: The concepts of gender identity and gestational age may be collected more than once during a study. Although these concepts are generally thought to belong in the Subject Characteristics (SC) domain, the SDTMIG v3.3 indicates that subject characteristics collected more than once should be represented in the Subject Status (SS) domain. However, representing gender identity in both SC and SS, depending on how often it is collected, conflicts with the principle that data should be consistently represented in only a single place. This issue was evaluated by the CDISC Global Governance Group, which decided that the assumptions of the SC domain should be modified to allow the representation of subject characteristics regardless of the number of times the characteristic is collected. This decision will be published in a future version of the SDTMIG. In this TAUG-HIV, Sections 3.2 (Representation of Gender Identity over Time) and 5.4 (Gestational Age) reflect this modeling decision.
-
Published gestational age controlled terminology: CDISC Controlled Terminology includes both RPTESTCD/RPTEST for EGESTAGE/Estimated Gestational Age and SCTESTCD/SCTEST for GSTABRTH/Gestational Age at Birth. Because gestational age is considered to be a subject characteristic of the infant, rather than a reproductive system finding, the published controlled terminology will be reviewed and updated. Section 5.4, Gestational Age, provides additional details.
Use of Device In Use (DU) to represent vaginal ring drug measurements:According to published controlled terminology the DU domain is "a domain for the findings for the values of measurements and settings that are intentionally set on a device when it is in use. These are characteristics that exist for the device, and have a specific setting for a use instance." However, there is ambiguity regarding whether or not the domain is intended to represent:
- a domain for the findings for the values of (measurements and settings) that are intentionally set on a device when it is in use, or
- a domain for the findings for the (values of measurements) and (settings that are intentionally set) on a device when it is in use.
The Device Team has agreed that the second interpretation is what was originally intended when creating the domain. The definition and intended use will be clarified in the next version of the SDMTIG-MD. Because of this, the HIV Team felt it was appropriate to represent the vaginal ring drug measurements within DU (Section 6.2.1, Vaginal Ring).
- Use of --RSDISC (Reason for Treatment Discontinuation) in the Device Exposure (DX) domain:In the vaginal ring example (Section 6.2.1, Vaginal Ring), the DXRSDISC variable is used to represent both the reason why each individual ring application ended and the overall reason for discontinuation of treatment with a vaginal ring.
- Representation of the date of a particular medical history episode: The variable MHEVDTYP specifies the aspect of the medical condition or event by which MHSTDTC and/or the MHENDTC is defined. Current controlled terminology values are "DIAGNOSIS", "EPISODE", "EXACERBATION", and "SYMPTOM ONSET". When the date is for an episode or exacerbation, which episode or exacerbation (e.g., first, most recent, worst) must be specified. It has not yet been decided whether this information should be included in the value of MHEVDTYP (e.g., giving a value such as "MOST RECENT EPISODE"), or in a separate variable. In Section 4.1, Baseline Menstrual History, Example 1, a separate NSV (MHCRNORD) has been used to represent the fact that the date collected was for the most recent menstrual period.
- Calculation of a single analysis value that is based on the data from more than 1 subject: The ADaM Basic Data Structure (BDS) class currently is designed to support subject-level analysis (as well as site-level analysis). However, grouping a number of subjects as an analysis unit has not been addressed. Example 1 in Section 8.3, Pregnancy Outcomes Analysis, proposes the variable USUBJGR1 (USUBJGRy as a generic variable) to allow for the grouping of subjects for analysis purposes. This allows for the calculation of a single analysis value that is based on the data from more than 1 subject. Because the ADOUTCOM dataset is summarized by family, based on the values across multiple subjects, a structure based on family and parameter is needed—which does not fit within the BDS definition. Therefore, it is proposed as an ADAM OTHER class of dataset.
2 Overview of HIV
Human immunodeficiency virus (HIV) causes a viral infection which, if not adequately controlled by treatment, may lead to acquired immunodeficiency syndrome (AIDS). HIV can weaken a person's immune system by reducing CD4+ T cells. AIDS is defined by the World Health Organization (WHO) as the occurrence of any 1 of more than 20 AIDS-defining clinical conditions, including opportunistic infections and HIV-related cancers.[1]The U.S. Centers for Disease Control and Prevention (CDC) definition is similar, but includes alternative criteria of a CD4+ cell count of fewer than 200 10^6/L or a CD4+ T cell percentage of total lymphocytes of less than 14%.[2] The 2 major types of HIV are HIV-1 and HIV-2. HIV-1 is the most common and most pathogenic. HIV-2 is less pathogenic and has been largely confined to Africa,although some cases have occurred in other parts of the world.
There are several populations that have a disproportionate burden of HIV. These groups include "men who have sex with men, transgender women in many settings and heterosexual men and women who have sexual partners with undiagnosed or untreated HIV infection".[3] Another population of interest is pregnant women with HIV. There are various approaches to preventing the mother-to-infant transmission of HIV. In HIV prevention studies involving pregnant women, infants will receive HIV testing and may receive antiretroviral therapy (ART) based on the mother's HIV status.
HIV treatment guidelines suggest a combination of antiretroviral (ARV) drugs to decrease the viral load below the levels of detection of sensitive HIV-RNA assays.[3,4] Antibody therapy may also be used to achieve this reduction in viral load. Current guidelines suggest lifelong treatment. Prevention of HIV in vulnerable populations includes the use of ARVs or treatment of potential sources (e.g., infected sexual partners, infected pregnant mothers to prevent perinatal transmission) prior to potential exposure to prevent transmission of HIV. Studies to develop vaccines for HIV have so far been unsuccessful. Vaccine trials are ongoing and may one day result in this being a viable option for prevention of HIV infection.[5,6]
Co-infections and complications of HIV-infected populations such as tuberculosis, hepatitis, and cardiovascular disease are also of interest. CDISC has published several therapeutic area user guides specifically for these indications. These guides can be found on the CDISC website (https://www.cdisc.org/standards/therapeutic-areas ).
3 Subject and Disease Characteristics
Information about subject and disease characteristics generally includes events and activities that have affected the subject prior to the study. In the TAUG-HIV, this includes:
3.1 Diagnosis of HIV
Diagnosing a subject with HIV requires an assay with a high degree of sensitivity and specificity. This is most easily achieved via a diagnostic algorithm that includes multiple redundant or complementary assays, as opposed to a single test.
The following concept map depicts a widely used algorithm for determining HIV status. Note that this algorithm does not apply to newborn infants of HIV-infected mothers.
Concept Map. HIV Diagnostic Algorithm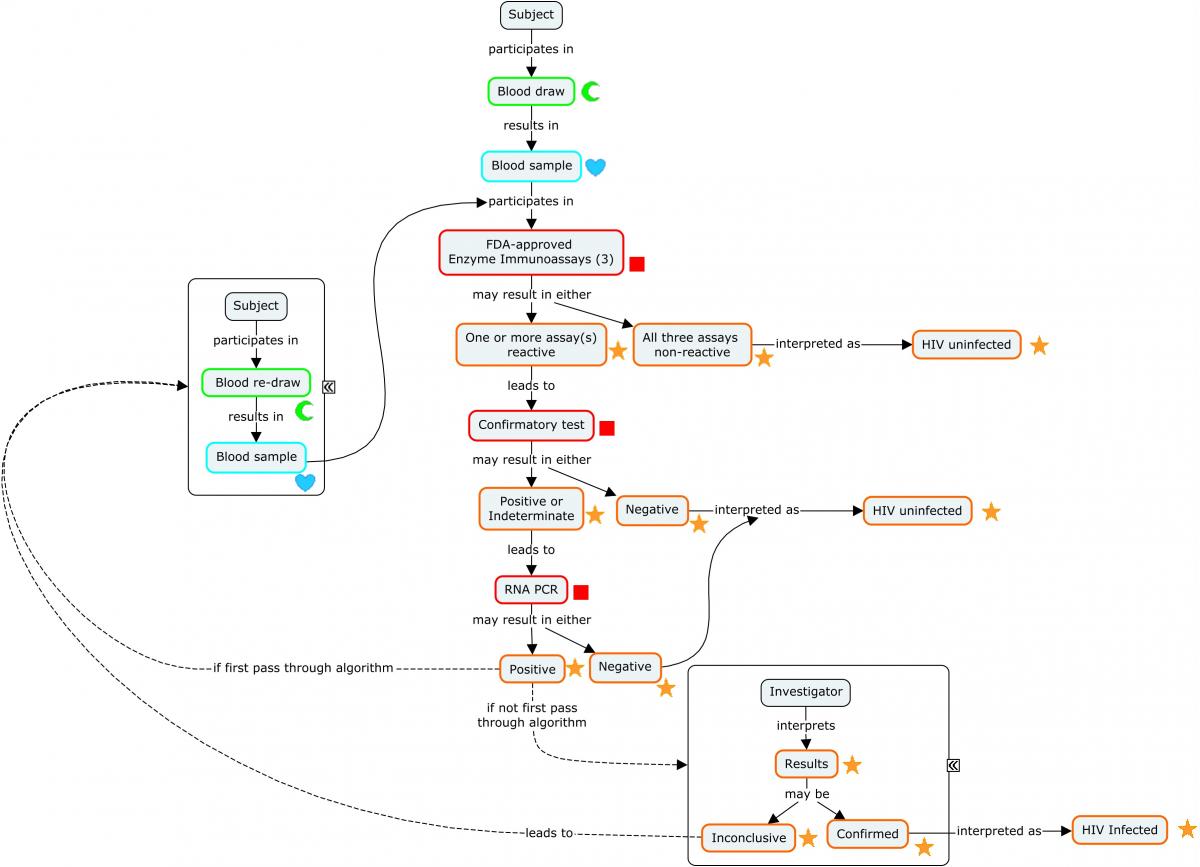
The following example illustrates a subject's progression through the diagnostic algorithm depicted in the concept map (second-stage confirmatory test results not shown).
Example
Because diagnosis of HIV involves a series of tests designed to detect antibodies to the virus, these findings are represented in the Immunogenicity Specimen Assessments (IS) domain. A final confirmatory test, which is designed to detect viral genetic material, is represented in the Microbiology Specimen (MB) domain. SPDEVID represents the device or kit used to obtain the individual results. The identifying details of these devices would be represented in the Device Identifiers (DI) dataset (example not shown).
The assays depicted in Rows 1-3 are antibody-only assays. Fourth-generation tests (combination tests that detect the presence of both HIV antibodies and HIV antigens) would be represented in the MB domain.
| Rows 1-9: | The combination of ISTESTCD / ISTEST and ISTSTDTL indicate that these tests are looking for the presence of the microbial-induced antibodies against the antigens identified in the non-standard variable ISBDAGNT. |
|---|---|
| Rows 1-3: | Show the subject was tested with 3 separate enzyme immunoassay tests for HIV and tested positive on 2 and indeterminate on the other. These tests detect the presence of antibodies against HIV-1 and HIV-2, but do not differentiate which virus antibodies were detected. |
| Rows 4-9: | Show the results from a confirmatory immunochromatographic assay that detects antibodies to specific antigens from both HIV-2 (Rows 4-5) and HIV-1 (Rows 6-9). Note that 2 distinct HIV-1 antibodies were detected (Rows 6 and 9). |
| Row 10: | Shows the subject is HIV-2 negative according to the assay identified in SPDEVID. The sponsor has chosen to group this interpretation record via ISGRPID to the 2 records from the assay that are relevant to HIV-2 antibody detection. |
| Row 11: | Shows the subject is HIV-1 positive according to the assay identified in SPDEVID. The sponsor has chosen to group this interpretation record via ISGRPID to the 4 records from the assay that are relevant to HIV-1 antibody detection. |
is.xpt
| Row | STUDYID | DOMAIN | USUBJID | ISSEQ | SPDEVID | ISGRPID | ISTESTCD | ISTEST | ISTSTDTL | ISORRES | ISSTRESC | ISSPEC | ISMETHOD | VISITNUM | VISIT | ISDTC | ISBDAGNT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC | IS | ABC-003 | 1 | EIA01 | MBAB | Binding Microbial-induced Antibody | DETECTION | DETECTED | DETECTED | BLOOD | EIA | 3 | WEEK 6 | 2017-05-22 | HIV-1/2 Antigen | ||
| 2 | ABC | IS | ABC-003 | 2 | EIA02 | MBAB | Binding Microbial-induced Antibody | DETECTION | INDETERMINATE | INDETERMINATE | BLOOD | EIA | 3 | WEEK 6 | 2017-05-22 | HIV-1/2 Antigen | ||
| 3 | ABC | IS | ABC-003 | 3 | EIA03 | MBAB | Binding Microbial-induced Antibody | DETECTION | DETECTED | DETECTED | BLOOD | EIA | 3 | WEEK 6 | 2017-05-22 | HIV-1/2 Antigen | ||
| 4 | ABC | IS | ABC-003 | 4 | 00105 | 1 | MBAB | Binding Microbial-induced Antibody | DETECTION | NOT DETECTED | NOT DETECTED | SERUM | IMMUNOCHROMATOGRAPHY | 3 | WEEK 6 | 2017-05-22 | HIV-2 GP36 Antigen | |
| 5 | ABC | IS | ABC-003 | 5 | 00105 | 1 | MBAB | Binding Microbial-induced Antibody | DETECTION | NOT DETECTED | NOT DETECTED | SERUM | IMMUNOCHROMATOGRAPHY | 3 | WEEK 6 | 2017-05-22 | HIV-2 GP140 Antigen | |
| 6 | ABC | IS | ABC-003 | 6 | 00105 | 2 | MBAB | Binding Microbial-induced Antibody | DETECTION | DETECTED | DETECTED | SERUM | IMMUNOCHROMATOGRAPHY | 3 | WEEK 6 | 2017-05-22 | HIV-1 P31 Antigen | |
| 7 | ABC | IS | ABC-003 | 7 | 00105 | 2 | MBAB | Binding Microbial-induced Antibody | DETECTION | NOT DETECTED | NOT DETECTED | SERUM | IMMUNOCHROMATOGRAPHY | 3 | WEEK 6 | 2017-05-22 | HIV-1 GP160 Antigen | |
| 8 | ABC | IS | ABC-003 | 8 | 00105 | 2 | MBAB | Binding Microbial-induced Antibody | DETECTION | NOT DETECTED | NOT DETECTED | SERUM | IMMUNOCHROMATOGRAPHY | 3 | WEEK 6 | 2017-05-22 | HIV-1 P24 Antigen | |
| 9 | ABC | IS | ABC-003 | 9 | 00105 | 2 | MBAB | Binding Microbial-induced Antibody | DETECTION | DETECTED | DETECTED | SERUM | IMMUNOCHROMATOGRAPHY | 3 | WEEK 6 | 2017-05-22 | HIV-1 GP41 Antigen | |
| 10 | ABC | IS | ABC-003 | 10 | 00105 | 1 | HIV2SR | HIV-2 Seroreactivity | INTERPRETATION | NEGATIVE | NEGATIVE | SERUM | IMMUNOCHROMATOGRAPHY | 3 | WEEK 6 | 2017-05-22 | ||
| 11 | ABC | IS | ABC-003 | 11 | 00105 | 2 | HIV1SR | HIV-1 Seroreactivity | INTERPRETATION | POSITIVE | POSITIVE | SERUM | IMMUNOCHROMATOGRAPHY | 3 | WEEK 6 | 2017-05-22 |
IS NSV Metadata
| Variable | Label | Type | Role | Origin |
|---|---|---|---|---|
| ISBDAGNT | Binding Agent | text | Non-Standard Record Qualifier | CRF |
The final confirmatory test detects the presence of viral RNA. Because this is not an antibody test, it is represented in MB rather than IS. This example shows that the subject was further tested for the presence of HIV-1 RNA using quantitative reverse transcriptase polymerase chain reaction. The target RNA was detected, confirming that the subject is HIV positive.
mb.xpt
| Row | STUDYID | DOMAIN | USUBJID | MBSEQ | MBTESTCD | MBTEST | MBTSTDTL | MBORRES | MBSTRESC | MBSPEC | MBMETHOD | VISITNUM | VISIT | MBDTC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC | MB | ABC-003 | 1 | HIV1RNA | HIV-1 RNA | DETECTION | DETECTED | DETECTED | BLOOD | QUANTITATIVE REVERSE TRANSCRIPTASE POLYMERASE CHAIN REACTION | 3 | WEEK 6 | 2017-05-22 |
Because this event occurred during the course of the study, it is represented in the Clinical Events (CE) domain.
ce.xpt
| Row | STUDYID | DOMAIN | USUBJID | CESEQ | CETERM | VISITNUM | VISIT | CEDTC |
|---|---|---|---|---|---|---|---|---|
| 1 | ABC | CE | ABC-003 | 1 | HIV test positive | 3 | WEEK 6 | 2017-05-22 |
There is no RELREC dataset included as part of this example. The sponsor could have used this dataset to create a link from the records in MB and IS, with the diagnosis record in CE. For more information on creating RELREC datastes, see SDTMIG Section 8.2, Relating Peer Records .
3.2 Representation of Gender Identity over Time
Transgender people are at high risk for HIV. Accurate capture and representation of gender identity in HIV prevention and treatment studies is important. Gender identity is the innermost concept of self as male, female, a blend of both, or neither. Gender is a spectrum, and not limited to just 2 possibilities. One's gender identity can be the same as the sex assigned at birth (cisgender) or different (transgender). A person may have a non-binary gender identity, meaning they do not identify strictly as male or female: They could identify as both, as neither, or as another gender entirely.[7,8,9]
Gender identity has traditionally been included in the Subject Characteristics (SC) domain. However, gender identity can be a fluid characteristic, especially in populations studied for HIV; thus it may be collected more than once during a study. A recent decision at CDISC allows subject characteristics collected as repeated measure to be included in SC. Further detail on this decision can be found in Section 1.4, Known Issues.
Because the terminology used to describe gender identity is evolving, CDISC has decided not to provide controlled terminology for potential values at this time. However, in order to populate the topic variable for SC consistently, a new SCTESTCD/SCTEST of "GENIDENT"/"Gender Identity" has been submitted to the CDISC Controlled Terminology Team.
In addition, the Tuberculosis Therapeutic Area User Guide v2.0 introduced the concept of "Sex Reported at Birth," which is also represented in the SC domain. The variable SEX in the Demographics (DM) domain is not well defined (sex of the subject) but is generally accepted to mean the self-reported sex of the subject. In support of this assumption, the CDASH Standard states that SEX is "the self-reported sex of the individual and/or is the clinician's assignment based on a physical examination. This is not to be confused with a genotypic determination of a subjects' chromosomally determined gender, but a less scientifically controlled method of visual determination that HL7 has defined as 'administrative sex.'" The concept of "Sex Reported at Birth" is intended to be used in cases where sex at birth differs from the subject's self-reported sex. However, sex and gender identity are complex issues that are only partially covered by the above concepts; further standards development work is required to adequately represent these concepts.
3.3 Risk Factors
Risk factors for contracting HIV include high-risk sexual behavior, intravenous drug use, prior sexually transmitted infections (STIs), and, for males, being uncircumcised.
Example 1 demonstrates a hypothetical CRF designed to capture the subject's history of HIV risk factors. The sponsor of this study chose to collect sexual behavior, sexually transmitted infections (STIs), intravenous drug use, and circumcision to assess the risk of HIV infection.
SDTM examples follow the aCRF.
This example refers to and makes use of the Environmental and Social Factors (ER) domain—a draft domain currently under discussion. As of the publication of this TAUG, the ER domain has no scheduled release date; the domain specification table can be found at https://wiki.cdisc.org/x/lIbIAQ.
Example
Annotated CRF: Risk Factors
This CRF collects information on e nvironmental and social risk factors. The specific STIs are collected on the Medical History CRF. In this example CRF, the following syntax was used to create denormalized CDASH variable names: SPONSOR DEFINED NAME_ROOT VARIABLE.
| Indicate if the subject has engaged in sexual contact without a condom with same-sex partners at any point in their life, by checking Yes or No. | Has the subject ever engaged in sexual contact without a condom with same-sex partners ?
UPSCSS_EROCCUR EROCCUR where ERTERM = "Sexual contact without a condom, same-sex partners" |
<From NY codelist> |
|---|---|---|
| Indicate if the subject has engaged in sexual contact without a condom with opposite-sex partners at any point in their life, by checking Yes or No. | Has the subject ever engaged in sexual contact without a condom with opposite-sex partners ?
UPSOP_EROCCUR EROCCUR where ERTERM = "Sexual contact without a condom, opposite-sex partners" |
<From NY codelist> |
| Indicate if the subject has ever been diagnosed with an STI. If Yes, complete the STI Medical History CRF. | Has the subject ever had a sexually transmitted infection (STI)?
PSTI_MHOCCUR MHOCCUR where MHTERM = "SEXUALLY TRANSMITTED INFECTIONS " |
<From NY codelist> |
| Indicate if the subject has been or is an IV drug user. | Has the subject ever been, or is now, an intravenous (IV) drug user?
IVU_EROCCUR EROCCUR where ERTERM = "Intravenous drug user" |
<From NY codelist> |
| Indicate if the subject has ever been an uncircumcised male since becoming sexually active. | Has the subject reported that he is, or that he has been, an uncircumcised male at any time since becoming sexually active?
UCM_EROCCUR EROCCUR where ERTERM = "Uncircumcised male" |
<From NY codelist> |
CRF Metadata
This Medical History CRF was used in this study to solicit the specific STI.
| Indicate if the subject has ever been diagnosed with gonorrhea. | Has the subject ever had gonorrhea?
GONORRHEA_MHOCCUR MHOCCUR where MHTERM = "GONORRHEA" |
<From NY codelist> |
|---|---|---|
| Record the start date of the medical event or condition. |
GONORRHEA_MHSTDAT MHSTDTC | _________________ |
| Indicate if the condition is ongoing at the time the history is collected. | Is the event ongoing at the time of collection of this history?
GONORRHEA_MHONGO MHENRTPT where MHENTPT = Date of Collection |
<From NY codelist> |
| Record the end date of the medical event or condition. |
GONORRHEA_MHENDAT MHENDTC | _________________ |
| Indicate if the subject has ever been diagnosed with chlamydia. | Has the subject ever had chlamydia?
CHLAMYDIA_MHOCCUR MHOCCUR where MHTERM = "CHLAMYDIA" |
<From NY codelist> |
| Record the start date of the medical event or condition. |
CHLAMYDIA_MHSTDAT MHSTDTC | _________________ |
| Indicate if the condition is ongoing. | Is the event ongoing at the time of collection of this history?
CHLAMYDIA_MHONGO MHENRTPT where MHENTPT = Date of Collection |
<From NY codelist> |
| Record the end date of the medical event or condition. |
CHLAMYDIA_MHENDAT MHENDTC | _________________ |
| Indicate if the subject has ever been diagnosed with genital warts. | Has the subject ever had genital warts?
GENITAL_WARTS_MHOCCUR MHOCCUR where MHTERM = "GENITAL WARTS" |
<From NY codelist> |
| Record the start date of the medical event or condition. |
GENITAL_WARTS_MHSTDAT MHSTDTC | _________________ |
| Indicate if the condition is ongoing. | Is the event ongoing at the time of collection of this history?
GENITAL_WARTS_MHONGO MHENRTPT where MHENTPT = Date of Collection |
<From NY codelist> |
| Record the end date of the medical event or condition. |
GENITAL_WARTS_MHENDAT MHENDTC | _________________ |
| Indicate if the subject has ever been diagnosed with genital herpes. | Has the subject ever had genital herpes?
GENITAL_HERPES_MHOCCUR MHOCCUR where MHTERM = "GENITAL HERPES" |
<From NY codelist> |
| Record the start date of the medical event or condition. |
GENITAL_HERPES_MHSTDAT MHSTDTC | _________________ |
| Indicate if the condition is ongoing at the time of collection of this history. | Is the event ongoing at the time of collection of this history?
GENITAL_HERPES_MHONGO MHENRTPT where MHENTPT = Date of Collection |
<From NY codelist> |
| Record the end date of the medical event or condition. |
GENITAL_HERPES_MHENDAT MHENDTC | _________________ |
| Indicate if the subject has ever been diagnosed with syphilis. | Has the subject ever had syphilis?
SYPHILIS_MHOCCUR MHOCCUR where MHTERM = "SYPHILIS" |
<From NY codelist> |
| Record the start date of the medical event or condition. |
SYPHILIS_MHSTDAT MHSTDTC | _________________ |
| Indicate if the condition is ongoing at the time of collection of this history. | Is the event ongoing at the time of collection of this history?
SYPHILIS_MHONGO MHENRTPT where MHENTPT = Date of Collection |
<From NY codelist> |
| Record the end date of the medical event or condition. |
SYPHILIS_MHENDAT MHENDTC | _________________ |
| Indicate if the subject has ever been diagnosed with hepatitis B. | Has the subject ever had hepatitis B?
HEPATITIS_B_MHOCCUR MHOCCUR where MHTERM = "HEPATITIS B" |
<From NY codelist> |
| Record the start date of the medical event or condition. |
HEPATITIS_B_MHSTDAT MHSTDTC | _________________ |
| Indicate if the condition is ongoing at the time of collection of this history. | Is the event ongoing at the time of collection of this history?
HEPATITIS_B_MHONGO MHENRTPT where MHENTPT = Date of Collection |
<From NY codelist> |
| Record the end date of the medical event or condition. |
HEPATITIS_B_MHENDAT MHENDTC | _________________ |
CRF Metadata
This example shows the results of a prespecified risk factors assessment for a subject enrolled in an HIV prevention study. This assessment is intended to be administered at the beginning of the study. Most of the items in the assessment pertain to any point in the subject's past, so EREVINTX is populated as "LIFETIME" for these items. One item pertains to the time since the subject became sexually active, and therefore has a value of "SINCE BECOMING SEXUALLY ACTIVE" in EREVINTX. In this example, the sponsor collected partial dates for the start date of the events .
Risk factors are represented in the Environmental and Social Factors (ER) domain. Because all of the risk factors queried for also exist as MedDRA terms, the sponsor chose to populate ERDECOD with the matching Preferred Term.
| Rows 1-2: | Show that the subject was asked about sexual contact without a condom. The subject indicated engagement in this risk-associated behavior with both same-sex and opposite-sex partners. |
|---|---|
| Row 3: | Shows that the subject has never been an intravenous drug user. |
| Row 4: | Shows that the subject has not been an uncircumcised male at any time since becoming sexually active. |
er.xpt
| Row | STUDYID | DOMAIN | USUBJID | ERSEQ | ERTERM | ERDECOD | ERCAT | ERPRESP | EROCCUR | ERDTC | EREVINTX |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC | ER | ABC-01-101 | 1 | Sexual contact without a condom, same-sex partners | High-risk sexual behavior | HIV RISK FACTORS | Y | Y | 2017-10-02 | LIFETIME |
| 2 | ABC | ER | ABC-01-101 | 2 | Sexual contact without a condom, opposite-sex partners | High-risk sexual behavior | HIV RISK FACTORS | Y | Y | 2017-10-02 | LIFETIME |
| 3 | ABC | ER | ABC-01-101 | 3 | Intravenous drug user | Drug abuser | HIV RISK FACTORS | Y | N | 2017-10-02 | LIFETIME |
| 4 | ABC | ER | ABC-01-101 | 4 | Uncircumcised male | Uncircumcised | HIV RISK FACTORS | Y | N | 2017-10-02 | SINCE BECOMING SEXUALLY ACTIVE |
In this study, the risk factors assessment CRF triggered the investigator to review the subject's medical history for the occurrence of selected specific STIs. The sponsor allowed partial dates to be provided for the start dates of any events. The investigators were required to indicate if any event was ongoing at the time of data collection; however, the actual end date was allowed to be missing. In the example below, MHCAT and MHSCAT are used to indicate that these items were collected as part of a risk factors assessment.
| Row 1: | Shows the subject responded "Yes" to the risk factors indicator question regarding past diagnosis of any STI. |
|---|---|
| Rows 2, 4: | Show that the subject had past diagnoses of gonorrhea and genital warts. MHENRTPT and MHENTPT are used to indicate that the event ended before the date of collection of the STI history but the end date was not known. |
| Rows 3, 5-7: | Show that the subject was asked about history of chlamydia, genital herpes, hepatitis B, and syphilis, but indicated never having contracted these diseases. |
mh.xpt
| Row | STUDYID | DOMAIN | USUBJID | MHSEQ | MHTERM | MHDECOD | MHCAT | MHSCAT | MHPRESP | MHOCCUR | MHDTC | MHSTDTC | MHENDTC | MHENRTPT | MHENTPT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC | MH | ABC-01-101 | 1 | SEXUALLY TRANSMITTED INFECTION | Sexually transmitted disease | HIV RISK FACTORS | HISTORY OF STI | Y | Y | 2017-10-02 | BEFORE | 2017-10-02 | ||
| 2 | ABC | MH | ABC-01-101 | 2 | GONORRHEA | Gonorrhoea | HIV RISK FACTORS | HISTORY OF STI | Y | Y | 2017-10-02 | 2016-09-09 | BEFORE | 2017-10-02 | |
| 3 | ABC | MH | ABC-01-101 | 3 | CHLAMYDIA | Chlamydial infection | HIV RISK FACTORS | HISTORY OF STI | Y | N | 2017-10-02 | ||||
| 4 | ABC | MH | ABC-01-101 | 4 | GENITAL WARTS | Anogenital warts | HIV RISK FACTORS | HISTORY OF STI | Y | Y | 2017-10-02 | 2013-04 | BEFORE | 2017-10-02 | |
| 5 | ABC | MH | ABC-01-101 | 5 | GENITAL HERPES | Genital herpes | HIV RISK FACTORS | HISTORY OF STI | Y | N | 2017-10-02 | ||||
| 6 | ABC | MH | ABC-01-101 | 6 | SYPHILIS | Syphilis | HIV RISK FACTORS | HISTORY OF STI | Y | N | 2017-10-02 | ||||
| 7 | ABC | MH | ABC-01-101 | 7 | HEPATITIS B | Hepatitis B | HIV RISK FACTORS | HISTORY OF STI | Y | N | 2017-10-02 |
3.4 Mode of Transmission
In HIV studies, it may be important to collect information about how the subject contracted the HIV virus (i.e., mode of disease transmission). At the broadest level, the mode of disease transmission can be categorized as either vertical transmission or horizontal transmission. Vertical transmission means that the virus is passed from mother to infant during pregnancy, birth, or breastfeeding. Horizontal transmission means that the virus is passed between individuals through activities such as sexual encounters and the sharing of IV needles.
As described below, HIV infections may be represented in either the Clinical Events (CE) domain or the Medical History (MH) domain, depending on the focus of the study. In either case, the mode of HIV transmission can be represented in the non-standard variable (NSV) --MODTR (Mode of Disease Transmission). If a study allows a subject to report more than 1 mode of transmission, --MODTR can be populated with "MULTIPLE". Additional NSVs can be added using the naming convention of --MODTR1 to --MODTRnto represent each mode of transmission reported. The level of granularity collected around the mode of transmission may vary by study. For example, a study may collect a category of mode of transmission such as "HORIZONTAL TRANSMISSION" or "VERTICAL TRANSMISSION". However, another study may collect a more granular mode of transmission that falls into 1 of these categories (e.g., "BLOOD TRANSFUSION", a type of horizontal transmission). Submission values can be found in the SDTM codelist, Mode of Disease Transmission (--MODTRN), and include:
| HORIZONTAL TRANSMISSION | VERTICAL TRANSMISSION |
|---|---|
| SAME-SEX SEXUAL CONTACT | INTRAUTERINE EXPOSURE |
| OPPOSITE-SEX SEXUAL CONTACT | BREASTFEEDING |
| SEXUAL CONTACT | LABOR AND DELIVERY |
| BLOOD TRANSFUSION | |
| INJECTION NEEDLE REUSE | |
| OCCUPATIONAL EXPOSURE |
Example
This example shows the mode of transmission collected as part of a prevention study where subjects may contract the HIV virus during the course of the study. In this scenario, the mode of transmission is represented as a non-standard variable of the CE domain. The study allowed subjects to report more than 1 mode of transmission. Mode of transmission may also be collected in a treatment study. In that case, because the subject is already infected with HIV at the beginning of the study, mode of transmission could be represented as a MH NSV.
ce.xpt
| Row | STUDYID | DOMAIN | USUBJID | CESEQ | CETERM | CEDECOD | CESTDTC | CEMODTR | CEMODTR1 | CEMODTR2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HIV-01 | CE | HIV-01-001 | 1 | HIV INFECTION | HIV infection | 2011-04-15 | MULTIPLE | OPPOSITE-SEX SEXUAL CONTACT | INJECTION NEEDLE REUSE | |
| 2 | HIV-01 | CE | HIV-01-002 | 1 | HIV INFECTION | HIV infection | 2010-07-08 | BLOOD TRANSFUSION | |||
| 3 | HIV-01 | CE | HIV-01-003 | 1 | HIV INFECTION | HIV infection | 2010-06-05 | MULTIPLE | INJECTION NEEDLE REUSE | SAME-SEX SEXUAL CONTACT | |
| 4 | HIV-01 | CE | HIV-01-004 | 1 | HIV INFECTION | HIV infection | 2009-02-02 | OCCUPATIONAL EXPOSURE | |||
| 5 | HIV-01 | CE | HIV-01-005 | 1 | HIV INFECTION | HIV infection | 2009-05-11 | UNKNOWN |
CE NSV Metadata
| Variable | Label | Type | Role | Origin |
|---|---|---|---|---|
| CEMODTR | Mode of Transmission | text | Non-Standard Record Qualifier | CRF |
| CEMODTR1 | Mode of Transmission 1 | text | Non-Standard Record Qualifier | CRF |
| CEMODTR2 | Mode of Transmission 2 | text | Non-Standard Record Qualifier | CRF |
4 Baseline Medical History and Physical Exams of Special Interest
This section contains examples and discussion on baseline menstrual history and pelvic examinations.
4.1 Baseline Menstrual History
HIV may cause more severe premenstrual symptoms for some women.[10] It is also possible that some women and girls living with or at risk for HIV, including those participating in clinical research for HIV/AIDS, do not menstruate. Reasons may include premenarche, menopause (natural or induced), long-acting contraceptives, and conditions that cause amenorrhea. Therefore, HIV/AIDS studies may need to collect more comprehensive data about the subject's gynecological assessments in order to place menstrual information within appropriate context.
Example 1 illustrates collecting information on premenstrual symptoms such as fatigue, cramps, headache, dysphoric disorder, pain, menstrual spotting, and diarrhea. Example 2 includes the number of days between menses and the range of usual bleeding days for the subject.
Example
This CRF was designed to collect menstrual history. General medical history was collected separately from this example. See Section 1.3, CDASH Metadata and Annotated CRFs, for explanation of annotations. In this example CRF, the following syntax was used to create denormalized CDASH variable names: <condition/event>_<root variable> (e.g., FATIGUE_MHOCCUR).
Baseline Menstrual History CRF
| Indicate if menses has occurred in the past 3 months by checking Yes or No. | In the past 3 months, has the subject had her menses?
MENSTRUAL_PERIOD_MHOCCUR MHOCCUR where MHTERM = "Menstrual Period" and MHEVLINT = "-P3M" |
<From NY codelist> |
|---|---|---|
| Indicate the bleeding extent of the menstrual flow by choosing Light, Moderate, or Heavy. | What was the bleeding extent of the most recent menses?
MHBLEXNT NSV.MHBLEXNT |
|
| Record the start date of the most recent menses. |
MENSTRUAL_PERIOD_MHSTDAT MHSTDTC | _________________ |
| Record the end date of the most recent menses. |
MENSTRUAL_PERIOD_MHENDAT MHENDTC | _________________ |
| Indicate if premenstrual symptoms have occurred by checking Yes or No. | Did the subject have premenstrual symptoms with the most recent menses?
PREMENSTRUAL_SYMPTOMS_MHOCCUR MHOCCUR where MHTERM = "Premenstrual Symptoms " and MHEVLINT = "-P3M" |
<From NY codelist> |
| Indicate if fatigue has occurred by checking Yes or No. | Did the subject have fatigue with the most recent menses?
FATIGUE_MHOCCUR MHOCCUR |
<From NY codelist> |
| Indicate if premenstrual cramps have occurred by checking Yes or No. | Did the subject have premenstrual cramps with the most recent menses?
PMCRAMPS_MHOCCUR MHOCCUR where MHTERM = "Premenstrual Cramps " |
<From NY codelist> |
| Indicate if headache has occurred by checking Yes or No. | Did the subject experience headache with the most recent menses?
HEADACHE_MHOCCUR MHOCCUR where MHTERM = "Headache " |
<From NY codelist> |
| Indicate if dysphoric disorder has occurred by checking Yes or No. | Did the subject have dysphoric disorder with the most recent menses?
DYSPHORIC_DISORDER_MHOCCUR MHOCCUR where MHTERM = "Dysphoric Disorder " |
<From NY codelist> |
| Indicate if pain has occurred by checking Yes or No. | Did the subject have pain with the most recent menses?
PAIN_MHOCCUR MHOCCUR where MHTERM = "Pain " |
<From NY codelist> |
| Indicate if spotting has occurred by checking Yes or No. | Did the subject experience menstrual spotting with the most recent menses?
MENSTRUAL_SPOTTING _MHOCCUR MHOCCUR where MHTERM = "Menstrual Spotting " |
<From NY codelist> |
| List other symptoms, 1 per row or page. |
OTHER_SYMPTOMS_MHTERM MHTERM where NSV.MHCRNORD = "MOST RECENT" | _________________ |
CRF Metadata
| Row 1: | Shows that the subject experienced a menstrual period within the last 3 months with the bleeding extent characterized as "moderate" (MHBLEXNT="MODERATE"). MHPRESP and MHOCCUR are equal to "Y" because the event has occurred and was prespecified on the CRF. |
|---|---|
| Rows 2-8: | Show that the subject experienced premenstrual symptoms, in general, as well as specific symptoms pre-specified on the CRF. MHSCAT="PREMENSTRUAL SYMPTOMS" and MHGRPID="1" and has been used to group the subject's premenstrual symptoms. The variables MHSTDTC and MHENDTC were not collected. The timing of these events is represented indirectly by the combination of the date of collection, MHDTC, with MHCRNORD="MOST RECENT". |
| Row 9: | Shows a record for a write-in premenstrual symptom recorded in the "Other, Specify" space. Because this event was not prespecified, MHOCCUR and MHPRESP are null. |
mh.xpt
| Row | STUDYID | DOMAIN | USUBJID | MHSEQ | MHGRPID | MHTERM | MHCAT | MHSCAT | MHPRESP | MHOCCUR | MHDTC | MHSTDTC | MHENDTC | MHEVLINT | MHCRNORD | MHBLEXNT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC | MH | ABC-001 | 1 | 1 | Menstrual Period | BASELINE MENSTRUAL HISTORY | Y | Y | 2016-06-01 | 2016-05-22 | 2016-05-27 | -P3M | MOST RECENT | MODERATE | ||
| 2 | ABC | MH | ABC-001 | 2 | 1 | Premenstrual Symptoms | BASELINE MENSTRUAL HISTORY | PREMENSTRUAL SYMPTOMS | Y | Y | 2016-06-01 | MOST RECENT | |||||
| 3 | ABC | MH | ABC-001 | 3 | 1 | Fatigue | BASELINE MENSTRUAL HISTORY | PREMENSTRUAL SYMPTOMS | Y | Y | 2016-06-01 | MOST RECENT | |||||
| 4 | ABC | MH | ABC-001 | 4 | 1 | Premenstrual Cramps | BASELINE MENSTRUAL HISTORY | PREMENSTRUAL SYMPTOMS | Y | Y | 2016-06-01 | MOST RECENT | |||||
| 5 | ABC | MH | ABC-001 | 5 | 1 | Headache | BASELINE MENSTRUAL HISTORY | PREMENSTRUAL SYMPTOMS | Y | Y | 2016-06-01 | MOST RECENT | |||||
| 6 | ABC | MH | ABC-001 | 6 | 1 | Dysphoric Disorder | BASELINE MENSTRUAL HISTORY | PREMENSTRUAL SYMPTOMS | Y | Y | 2016-06-01 | MOST RECENT | |||||
| 7 | ABC | MH | ABC-001 | 7 | 1 | Pain | BASELINE MENSTRUAL HISTORY | PREMENSTRUAL SYMPTOMS | Y | Y | 2016-06-01 | MOST RECENT | |||||
| 8 | ABC | MH | ABC-001 | 8 | 1 | Menstrual Spotting | BASELINE MENSTRUAL HISTORY | PREMENSTRUAL SYMPTOMS | Y | Y | 2016-06-01 | MOST RECENT | |||||
| 9 | ABC | MH | ABC-001 | 9 | 1 | Diarrhea | BASELINE MENSTRUAL HISTORY | PREMENSTRUAL SYMPTOMS | 2016-06-01 | MOST RECENT |
MH NSV Metadata
| Variable | Label | Type | Role | Origin |
|---|---|---|---|---|
| MHCRNORD | Chronological Order | text | Record Qualifier | CRF |
| MHBLEXNT | Bleeding Extent | text | Non-Standard Record Qualifier | CRF |
Example
HIV may alter the duration, frequency, or bleeding extent of a woman's menstrual cycle. It may be important to collect these data during an HIV study. See Section 1.3, CDASH Metadata and Annotated CRFs, for explanation of annotations. In this example CRF, the following syntax was used to create denormalized CDASH variable names: <condition/event>_<root variable> (e.g., MENARAGE_RPORRES ).
Baseline Menstrual History CRF 2
| Record the date when the menstrual history was collected, using this format: DD-MON-YYYY. |
RPDAT RPDTC | _________________ |
|---|---|---|
| Record the menarche age for the subject. |
MENARAGE_RPORRES RPORRES where RPTESTCD = "MENARAGE" | _____ |
| Record the subject's u sual number of days between menses . |
NUMBTWMN _RPORRES RPORRES where RPTESTCD = "NUMBTWMN" | _____ |
| Record the usual minimum number of bleeding days for the subject. |
NUMBLMIN _RPORRES RPORRES where RPTESTCD = " NUMBLMIN " | _____ |
| Record the usual maximum number of bleeding days for the subject. |
NUMBLMAX _RPORRES RPORRES where RPTESTCD = " NUMBLMAX " | _____ |
CRF Metadata
| Row 1: | Shows that the subject began menstruating at the age of 13 years . |
|---|---|
| Rows 2-4: | Show various details regarding the subject's usual menstrual cycle, including number of days between menses and the range of the usual bleeding days. |
rp.xpt
| Row | STUDYID | DOMAIN | USUBJID | RPSEQ | RPTESTCD | RPTEST | RPCAT | RPORRES | RPORRESU | RPSTRESC | RPSTRESN | RPSTRESU | VISITNUM | VISIT | RPDTC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC | RP | ABC-001 | 1 | MENARAGE | Menarche Age | BASELINE MENSTRUAL HISTORY | 13 | YEARS | 13 | 13 | YEARS | 1 | SCREENING | 2016-06-01 |
| 2 | ABC | RP | ABC-001 | 2 | NUMBTWMN | Usual Number of Days Between Menses | BASELINE MENSTRUAL HISTORY | 28 | DAYS | 28 | 28 | DAYS | 1 | SCREENING | 2016-06-01 |
| 3 | ABC | RP | ABC-001 | 3 | NUMBLMIN | Usual Number of Bleeding Days, Min | BASELINE MENSTRUAL HISTORY | 4 | DAYS | 4 | 4 | DAYS | 1 | SCREENING | 2016-06-01 |
| 4 | ABC | RP | ABC-001 | 4 | NUMBLMAX | Usual Number of Bleeding Days, Max | BASELINE MENSTRUAL HISTORY | 7 | DAYS | 7 | 7 | DAYS | 1 | SCREENING | 2016-06-01 |
4.2 Pelvic Examination
During a pelvic examination, an investigator may check for prespecified abnormalities. When this occurs, the prespecified finding of interest is represented in RPTESTCD/RPTEST. The value in the variable RPORRES indicates whether the prespecified finding of interest was observed. Example 1 shows the representation of data from a prespecified pelvic exam for a single subject.
Example
In this example, the mother was a study subject. If the mother were an associated person, the data would be represented in the APMH domain and the dataset would include APID, RSUBJID, and SREL, rather than USUBID.
rp.xpt
| Row | STUDYID | DOMAIN | USUBJID | RPSEQ | RPTESTCD | RPTEST | RPCAT | RPORRES | RPSTRESC | RPLOC | VISITNUM | VISIT | RPDTC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC | RP | ABC-001 | 1 | EDEMAIND | Edema Indicator | PELVIC EXAM FINDINGS | Y | Y | VULVA | 1 | SCREENING | 2016-02-08 |
| 2 | ABC | RP | ABC-001 | 2 | ERYTHIND | Erythema Indicator | PELVIC EXAM FINDINGS | Y | Y | VULVA | 1 | SCREENING | 2016-02-08 |
| 3 | ABC | RP | ABC-001 | 3 | RASHIND | Rash Indicator | PELVIC EXAM FINDINGS | N | N | VULVA | 1 | SCREENING | 2016-02-08 |
| 4 | ABC | RP | ABC-001 | 4 | TENDIND | Tenderness Indicator | PELVIC EXAM FINDINGS | Y | Y | VULVA | 1 | SCREENING | 2016-02-08 |
| 5 | ABC | RP | ABC-001 | 5 | SKGLAIND | Skene's Gland Abnormality Indicator | PELVIC EXAM FINDINGS | N | N | VULVA | 1 | SCREENING | 2016-02-08 |
| 6 | ABC | RP | ABC-001 | 6 | BTHGLIND | Bartholin's Gland Abnormality Indicator | PELVIC EXAM FINDINGS | N | N | VULVA | 1 | SCREENING | 2016-02-08 |
| 7 | ABC | RP | ABC-001 | 7 | ULCERIND | Ulcer Indicator | PELVIC EXAM FINDINGS | Y | Y | VULVA | 1 | SCREENING | 2016-02-08 |
| 8 | ABC | RP | ABC-001 | 8 | BLISTIND | Blister Indicator | PELVIC EXAM FINDINGS | Y | Y | VULVA | 1 | SCREENING | 2016-02-08 |
| 9 | ABC | RP | ABC-001 | 9 | PUSTIND | Pustule Indicator | PELVIC EXAM FINDINGS | N | N | VULVA | 1 | SCREENING | 2016-02-08 |
| 10 | ABC | RP | ABC-001 | 10 | PEELIND | Peeling Indicator | PELVIC EXAM FINDINGS | Y | Y | VULVA | 1 | SCREENING | 2016-02-08 |
| 11 | ABC | RP | ABC-001 | 11 | ECCHYIND | Ecchymosis Indicator | PELVIC EXAM FINDINGS | N | N | VULVA | 1 | SCREENING | 2016-02-08 |
| 12 | ABC | RP | ABC-001 | 12 | EDEMAIND | Edema Indicator | PELVIC EXAM FINDINGS | Y | Y | CERVIX UTERI | 1 | SCREENING | 2016-02-08 |
| 13 | ABC | RP | ABC-001 | 13 | ERYTHIND | Erythema Indicator | PELVIC EXAM FINDINGS | Y | Y | CERVIX UTERI | 1 | SCREENING | 2016-02-08 |
| 14 | ABC | RP | ABC-001 | 14 | MASSIND | Mass Indicator | PELVIC EXAM FINDINGS | N | N | CERVIX UTERI | 1 | SCREENING | 2016-02-08 |
| 15 | ABC | RP | ABC-001 | 15 | MTENDIND | Motion Tenderness Indicator | PELVIC EXAM FINDINGS | N | N | CERVIX UTERI | 1 | SCREENING | 2016-02-08 |
| 16 | ABC | RP | ABC-001 | 16 | DSCHGIND | Discharge Indicator | PELVIC EXAM FINDINGS | Y | Y | CERVIX UTERI | 1 | SCREENING | 2016-02-08 |
| 17 | ABC | RP | ABC-001 | 17 | ULCERIND | Ulcer Indicator | PELVIC EXAM FINDINGS | N | N | CERVIX UTERI | 1 | SCREENING | 2016-02-08 |
| 18 | ABC | RP | ABC-001 | 18 | BLISTIND | Blister Indicator | PELVIC EXAM FINDINGS | Y | Y | CERVIX UTERI | 1 | SCREENING | 2016-02-08 |
| 19 | ABC | RP | ABC-001 | 19 | PUSTIND | Pustule Indicator | PELVIC EXAM FINDINGS | N | N | CERVIX UTERI | 1 | SCREENING | 2016-02-08 |
| 20 | ABC | RP | ABC-001 | 20 | PEELIND | Peeling Indicator | PELVIC EXAM FINDINGS | Y | Y | CERVIX UTERI | 1 | SCREENING | 2016-02-08 |
| 21 | ABC | RP | ABC-001 | 21 | ECCHYIND | Ecchymosis Indicator | PELVIC EXAM FINDINGS | N | N | CERVIX UTERI | 1 | SCREENING | 2016-02-08 |
| 22 | ABC | RP | ABC-001 | 22 | BLEEDIND | Abnormal Bleeding Indicator | PELVIC EXAM FINDINGS | N | N | FEMALE GENITALIA | 1 | SCREENING | 2016-02-08 |
5 Mother-Infant Pairs in HIV Studies
Given the high burden of HIV infection among women of childbearing potential, particularly in low- and middle-income settings, the study of HIV treatment and prevention in HIV-infected pregnant women and their infants is of high priority. When a subject is pregnant or becomes pregnant, it is important to track pregnancy outcomes and it may be necessary to collect both prenatal and postnatal data on the infant pertaining to overall health and HIV status.
In studies that collect perinatal data, the term used to describe the child changes over time, from fetus to newborn (or neonate) to infant. Within particular examples where the child is at a particular stage, the appropriate term will be used, according to definitions in Appendix B, Glossary and Abbreviations. Statements in this document that apply to multiple stages use the term infant.
In SDTM-based datasets, data about persons who are not study subjects (associated persons) are represented differently from data about study subjects. This means that representation of data about mothers and infants depends on whether they are study subjects or associated persons—which, in turn, depends on the study design and protocol. This section covers the representation of data collected about mothers and infants and the representation of the relationship between mother and infant in studies where one or the other or both are study subjects.
5.1 Studies Involving Mother-Infant Data
The roles that mothers and infants play in a study determine how pregnancy and perinatal data are represented in the SDTM. This TAUG-HIV includes examples that illustrate data in one of 3 categories of study: those that enroll women of childbearing potential, studies of mothers and infants, and studies of infants.
Studies that Enroll Women of Childbearing Potential
If a woman of childbearing potential becomes pregnant during a study, the protocol may call for collection of data about the pregnancy and minimal data about the infant(s). In such a study, the infant is unlikely to be treated as a study subject, but rather as an associated person or a non-study subject. Any data about the infant would be represented in an associated persons (AP) dataset, as described in the SDTMIG-AP.
An associated person is given an associated person identifier (APID), rather than a unique subject identifier (USUBJID). AP datasets include the APID, the USUBJID of the study subject with whom they are associated (RSUBJID), and a variable (SREL) that describes the relationship of the associated person to the study subject. SREL would be a value such as "CHILD, BIOLOGICAL" for infants of mothers in a study in this category.
Studies of Mothers and Infants
Some studies enroll pregnant women and treat a mother and her infant(s) as study subjects. In this case, each person has a unique subject identifier (USUBJID). Relationships between study subjects can be represented in the Related Subjects dataset (RELSUB).
Studies of Infants
There are occasional studies of neonates which collect data about mothers, but in which the mothers are associated persons, rather than study subjects. Maternal data would be represented in AP domains.
5.2 Identifiers and Relationships for Mother-Infant Pairs
The mother-infant relationship is represented in the SDTM in a RELSUB dataset if both mother and the infant are study subjects, and in an Associated Persons Related Subjects (APRELSUB) dataset if only 1 is a study subject. The APRELSUB dataset is not necessary unless there are associated persons who have either relationships with multiple study subjects or multiple relationships with the same study subject. However, the APRELSUB dataset can be helpful even when not required, as a single dataset that shows all relationships of associated persons to study subjects.
Because multiple gestations during a single pregnancy increase the complexity of data modeling, many of the examples shown in this section are based on a mother who is pregnant with or has given birth to twins. In the examples for scenarios in which the mother is a study subject, the infant identifiers are shown as the mother's study identifier with the letter "A" or "B" appended. In the scenario where the mother is an associated person, the mother's identifier is based on the identifier of an infant.
When the mother is the study subject and the infant is an associated person, the relationship of the infant to the mother is represented in each record in an AP dataset, since AP datasets include the variables RSUBJID (Related Subject Identifier) and SREL (Subject Relationship). The relationship can also be represented in the APRELSUB dataset. Relationships are described as the relationship of the associated person to the study subject, so only 1 record is required for each relationship.
Example
This example is for a study in which mothers were study subjects and infants were associated persons. In this example, a mother gave birth to twins and the sponsor assigned associated person identifiers (APIDs) to infants by appending a letter ("A" and "B" in this scenario) to the mother's ID. Here, the relationship between an associated person and a study subject is represented in APRELSUB.
aprelsub.xpt
| Row | STUDYID | APID | RSUBJID | SREL |
|---|---|---|---|---|
| 1 | ABC-1 | 101A | 101 | SON, BIOLOGICAL |
| 2 | ABC-1 | 101B | 101 | DAUGHTER, BIOLOGICAL |
When the infant and the mother are both study subjects, they are each assigned a unique subject identifier (USUBJID) and the relationship is defined in the RELSUB dataset. The RELSUB dataset always shows each relationship between 2 study subjects using 2 records, since relationships (represented in SREL) are generally not symmetrical.
Example
This example is for a study in which both mothers and infants were study subjects. In this example, a mother gave birth to twins and the sponsor assigned unique subject identifiers (USUBJID) to infants by appending a letter ("A" and "B" in this scenario) to the mother's unique subject identifier (USUBJID).
| Rows 1-2: | Show the relationships between the mother and the first infant. |
|---|---|
| Rows 3-4: | Show the relationships between the mother and the second infant. |
relsub.xpt
| Row | STUDYID | USUBJID | RSUBJID | SREL |
|---|---|---|---|---|
| 1 | ABC-2 | 101 | 101A | MOTHER, BIOLOGICAL |
| 2 | ABC-2 | 101A | 101 | SON, BIOLOGICAL |
| 3 | ABC-2 | 101 | 101B | MOTHER, BIOLOGICAL |
| 4 | ABC-2 | 101B | 101 | DAUGHTER, BIOLOGICAL |
When the infant is the study subject and the mother is an associated person, the relationship can be represented in the APRELSUB dataset. Relationships are described as the relationship of the associated person to the study subject, so only 1 record is required for each relationship.
Example
Infants were study subjects in this study; their mothers were associated persons. In this example, twins with the study identifiers "101" and "102" were study subjects. Their mother was an associated person. Because she was related to 2 different study subjects, the APRELSUB dataset was required to represent her relationships to study subjects. In this study, the sponsor assigned identifiers to mothers by appending "M" to infant study identifiers. In this case, the sponsor chose the identifier "101M" for the mother based on the identifier of 1 of the twins.
aprelsub.xpt
| Row | STUDYID | APID | RSUBJID | SREL |
|---|---|---|---|---|
| 1 | ABC-3 | 101M | 101 | MOTHER, BIOLOGICAL |
| 2 | ABC-3 | 101M | 102 | MOTHER, BIOLOGICAL |
5.3 Representation of Pregnancy-related Events
Pregnancies are represented as events. A woman could become pregnant more than once during a long study. In this TAUG-HIV, --LNKID is used as a pregnancy identifier, to link data within and across domains for the same pregnancy.
The amount of data collected about pregnancy outcome is likely to depend on the time at which the pregnancy ends. In this guide, 20 weeks has been used as the dividing line between early and late pregnancy outcomes. The 20-week division is derived from the definitions of spontaneous abortion (also known as miscarriage) as something that happens before 20 weeks, and late fetal death as something that happens after 20 weeks. ( Complications of pregnancy are covered in Section 5.8, Routine Data for Mother-Infant Pairs.)
In this TAUG, pregnancy-related data are modeled as data about the mother, which means that pregnancies with multiple infants complicate the modeling when the outcome and the way in which the fetuses separate from the mother can differ. It is assumed that in an early pregnancy termination (before 20 weeks), the pregnancy end is the same for all fetuses, so separate records for multiple fetuses are not needed. A non-standard qualifier, Fetus/Infant Identifier (–FTINID), has been used to distinguish between multiple maternal records for pregnancies that end after 20 weeks. Although the SDTM includes the variable FETUSID, the variable is not intended for use in human clinical trials, and was intended only to distinguish between records for a particular test, not to be consistent across tests. The --FTINID in this guide is intended to identify fetuses consistently across records at a particular time (e.g., at birth), but not necessarily across times (e.g., across ultrasounds at various times during pregnancy).
The outcome of a pregnancy is modeled as an attribute of the mother's pregnancy as "Early pregnancy termination", "Late fetal death", "Stillbirth", or "Live birth", although there is an outcome for each fetus. This combines survival of the fetus with the stage at which a non-surviving fetus died.
The way in which a fetus is separated from the mother may be natural, as in a miscarriage or an unassisted vaginal delivery, or by means of an intervention such as an induced abortion or a Cesarean delivery. In the examples below, all ways in which a pregnancy ends are modeled as events, with the rationale that all (with the possible exception of live birth) are things which might be reported as adverse events. Events which can also be considered interventions present a choice for the sponsor, similar to the choice of whether to create a Procedures (PR) domain record for procedures whose main purpose is to gather data reported in findings domains. In the examples below, surgical abortion, surgical termination of an ectopic pregnancy, Cesarean delivery, forceps-assisted vaginal delivery, and vacuum-assisted vaginal delivery are modeled as procedures; medication-induced abortion and termination of an ectopic pregnancy are modeled as a concomitant medication administrations. Some of the data collected in the examples below, such as the urgency of a cesarean delivery or the medication used to induce pregnancy termination, are interventions data, and are best represented in an interventions record or a supplemental qualifier to an interventions record. If such data are not collected, the sponsor can decide whether to create interventions records for interventions that separate a fetus from its mother.
This modeling is illustrated in the following concept map, which includes SDTM domains and selected variables.
Concept Map. Pregnancy-Related Data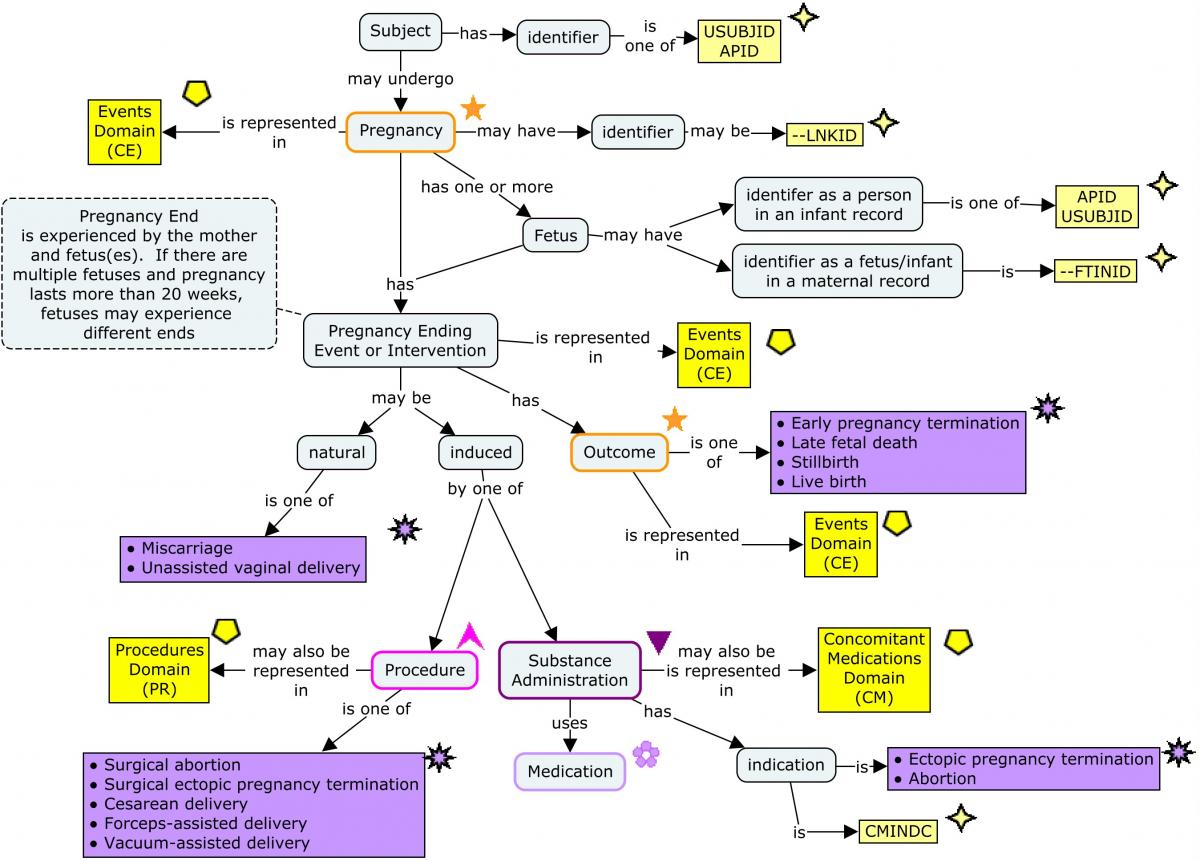
The following examples show pregnancy-related data as it would appear in the categories of studies described in Section 5.2, Identifiers and Relationships for Mother-Infant Pairs.
Example
This is an example in which women are study subjects, and any data about their infants are represented in Associated Persons (AP) datasets.
This example CRF shows data collected about a pregnancy. Different data are collected for pregnancies that lasted less than and more than 20 weeks. In this example, the 2 identifier variables --LNKID and CEFTINID are shown as though they were entered, but in an electronic data-capture system, system variables that distinguish between instances of a form could be used as the basis for these identifier variables.
Pregnancy Outcome aCRF
Complete for each pregnancy.
|
CECAT, CMCAT, PRCAT Pre-populated | PREGNANCY-RELATED EVENTS | |
|
CELNKID, CEGRPID CELNKID, CMLNKID, PRLNKID, CEGRPID | _____ | |
|
CESTDAT CESTDTC | _ _ / _ _ _ / _ _ _ _ | |
|
CEENDAT CEENDTC, CMSTDTC, PRSTDTC | _ _ / _ _ _ / _ _ _ _ |
CRF Metadata
If the pregnancy lasted less than 20 weeks, fill in this form.
| Select the most appropriate response. | What is the clinical event term?
END_CETERM CETERM, CMINDC, PRTRT |
|
|---|---|---|
|
END_CMTRT CMTRT | _________________ |
CRF Metadata
If the pregnancy lasted 20 weeks or more, provide the number of fetuses in the pregnancy.
|
CEFETNUM | _____ |
CRF Metadata
If the pregnancy lasted 20 weeks or more, fill in this form for each gestation.
|
CEFTINID CEFTINID, PRFTINID | _____ | |
| Select the most appropriate response. | What is the clinical event term?
OUTCOME_CETERM CETERM |
|
| Select the most appropriate response. | If still birth or live birth, what was the setting?
OUTCOME_CESETTNG CESETTNG |
<From SETTING codelist> |
|
OUTCOME_CESETTOTH CESETTING | _________________ | |
| Select the most appropriate response. | What was the type of delivery?
DELIVERY_CETERM CETERM, PRTRT |
|
| Select the most appropriate response. | If forceps- or vacuum-assisted vaginal delivery, what was the location in the birth canal?
BCLOC_PRTRT PRBCLOC |
|
| Select the most appropriate response. | If cesarean delivery, what was the urgency?
PRURGNCY |
<From PRURGNCY codelist> |
CRF Metadata
The data collected on this CRF would be represented in up to 3 datasets:
- Clinical Events (CE) for pregnancy, pregnancy outcome, and natural ends to pregnancy;
- PR for procedures which ended pregnancy; and
- Concomitant/Prior Medications (CM) for medication-induced pregnancy terminations.
In this example, the non-standard identifier CEFTINID was used to distinguish between infants in a multiple-gestation pregnancy. The CESPID variable was considered for this purpose but was not used, because --SPID is a sponsor-defined variable for which a sponsor may have other uses.
The CE dataset has at least 2 records for each pregnancy: a record for the pregnancy and a record for pregnancy outcome. Pregnancies of more than 20 weeks with more than 1 fetus have a pregnancy outcome record for each fetus. Natural ends of pregnancy (e.g., miscarriage, unassisted vaginal birth) are also represented in the CE domain.
| Rows 1-6: | Show 2 pregnancies for the same subject, distinguished by CELNKID="PREG1" or "PREG2". Both pregnancies had outcomes of "Early pregnancy termination", both by miscarriage. |
|---|---|
| Rows 7-8: | Show a pregnancy that ended in late fetal death. |
| Rows 9-13: | Show events related to a pregnancy with 2 fetuses. The fetus identified by --FTINID="1" experienced unassisted vaginal delivery, and was stillborn in a vehicle. The fetus identified by --FTINID="2" was born alive in a hospital and delivered by Cesarean delivery. The sponsor also represented the cesarean delivery in a PR domain record linked to records in CE using CELNKID and PRLNKID. |
| Rows 14-16: | Show a pregnancy that terminated early. The pregnancy ended in a procedure which was also represented in the PR domain, linked to the CE records using CELNKID and PRLNKID. |
| Rows 17-19: | Show a singleton pregnancy that resulted in a live birth in a hospital by forceps-assisted vaginal delivery. The forceps-assisted vaginal delivery was also represented in the PR domain, linked to the CE records using CELNKID and PRLNKID. |
| Rows 20-22: | Show another pregnancy that ended early. In this case, the pregnancy was ended by administration of a medication, represented in the CM domain and linked to CE records using CMLNKID. |
ce.xpt
| Row | STUDYID | DOMAIN | USUBJID | CESEQ | CELNKID | CETERM | CECAT | CESCAT | CESTDTC | CEENDTC | CEFETNUM | CEFTINID | CESETTNG | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC-123 | CE | 101 | 1 | PREG1 | Pregnancy | PREGNANCY-RELATED EVENTS | 2017-01-10 | 2017-05-15 | |||||
| 2 | ABC-123 | CE | 101 | 2 | PREG1 | Early pregnancy termination | PREGNANCY-RELATED EVENTS | PREGNANCY OUTCOME | 2017-05-15 | |||||
| 3 | ABC-123 | CE | 101 | 3 | PREG1 | Miscarriage | PREGNANCY-RELATED EVENTS | 2017-05-15 | ||||||
| 4 | ABC-123 | CE | 101 | 4 | PREG2 | Pregnancy | PREGNANCY-RELATED EVENTS | 2017-07-04 | 2017-09-02 | |||||
| 5 | ABC-123 | CE | 101 | 5 | PREG2 | Early pregnancy termination | PREGNANCY-RELATED EVENTS | PREGNANCY OUTCOME | 2017-05-15 | |||||
| 6 | ABC-123 | CE | 101 | 6 | PREG2 | Miscarriage | PREGNANCY-RELATED EVENTS | 2017-09-02 | ||||||
| 7 | ABC-123 | CE | 102 | 1 | PREG1 | Pregnancy | PREGNANCY-RELATED EVENTS | 2017-11-23 | 2018-06-18 | 1 | ||||
| 8 | ABC-123 | CE | 102 | 2 | PREG1 | Late fetal death | PREGNANCY-RELATED EVENTS | PREGNANCY OUTCOME | 2017-06-18 | |||||
| 9 | ABC-123 | CE | 103 | 1 | PREG1 | Pregnancy | PREGNANCY-RELATED EVENTS | 2017-01-10 | 2017-10-25 | 2 | ||||
| 10 | ABC-123 | CE | 103 | 2 | PREG1 | Stillbirth | PREGNANCY-RELATED EVENTS | PREGNANCY OUTCOME | 2017-10-25 | 1 | VEHICLE | |||
| 11 | ABC-123 | CE | 103 | 3 | PREG1 | Unassisted vaginal delivery | PREGNANCY-RELATED EVENTS | 2017-10-25 | 1 | |||||
| 12 | ABC-123 | CE | 103 | 4 | PREG1 | Live birth | PREGNANCY-RELATED EVENTS | PREGNANCY OUTCOME | 2017-10-25 | 2 | HOSPITAL | |||
| 13 | ABC-123 | CE | 103 | 5 | PREG1 | Cesarean delivery | PREGNANCY-RELATED EVENTS | 2017-10-25 | 2 | |||||
| 14 | ABC-123 | CE | 104 | 1 | PREG1 | Pregnancy | PREGNANCY-RELATED EVENTS | 2017-02-27 | 2017-05-15 | 1 | ||||
| 15 | ABC-123 | CE | 104 | 2 | PREG1 | Early pregnancy termination | PREGNANCY-RELATED EVENTS | PREGNANCY OUTCOME | 2017-05-15 | 1 | ||||
| 16 | ABC-123 | CE | 104 | 3 | PREG1 | Surgical ectopic pregnancy termination | PREGNANCY-RELATED EVENTS | 2017-05-15 | 1 | |||||
| 17 | ABC-123 | CE | 105 | 1 | PREG1 | Pregnancy | PREGNANCY-RELATED EVENTS | 2017-09-23 | 2018-06-01 | 1 | ||||
| 18 | ABC-123 | CE | 105 | 2 | PREG1 | Live birth | PREGNANCY-RELATED EVENTS | PREGNANCY OUTCOME | 2017-06-01 | 1 | HOSPITAL | |||
| 19 | ABC-123 | CE | 105 | 3 | PREG1 | Forceps-assisted delivery | PREGNANCY-RELATED EVENTS | 2018-06-01 | 1 | |||||
| 20 | ABC=23 | CE | 106 | 1 | PREG1 | Pregnancy | PREGNANCY-RELATED EVENTS | 2017-06-22 | 2017-08-14 | |||||
| 21 | ABC=23 | CE | 106 | 2 | PREG1 | Early pregnancy termination | PREGNANCY-RELATED EVENTS | PREGNANCY OUTCOME | 2017-08-14 | |||||
| 22 | ABC=23 | CE | 106 | 3 | PREG1 | Medication-induced ectopic pregnancy termination | PREGNANCY-RELATED EVENTS | PREGNANCY OUTCOME | 2017-08-14 |
CE NSV Metadata
| Variable | Label | Type | Role | Codelist | Origin |
|---|---|---|---|---|---|
| CEFETNUM | Number of Fetuses/Infants in Pregnancy | integer | Non-Standard Record Qualifier | CRF | |
| CEFTINID | Fetus/Infant Identifier | integer | Non-Standard Identifier | CRF | |
| CESETTNG | Event Setting | text | Non-Standard Record Qualifier | SETTING | CRF |
The termination of a subject's ectopic pregnancy by means of the administration of a medication was represented in the CM domain, with "Ectopic pregnancy termination" as the indication.
cm.xpt
| Row | STUDYID | DOMAIN | USUBJID | CMSEQ | CMLNKID | CMTRT | CMINDC | CMDOSFRQ | CMSTDTC | CMSTDY |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC-103 | CM | 106 | 1 | PREG1 | Methotrexate | Ectopic pregnancy termination | ONCE | 2017-08-14 | 62 |
The sponsor represented termination of pregnancies by surgical means with records in the PR domain.
| Row 1: | Shows that for the infant identified with --FTINID="2", pregnancy ended in cesarean delivery. The urgency of this procedure was represented with the non-standard variable PRURGNCY. |
|---|---|
| Row 2: | Shows an ectopic pregnancy that was terminated surgically. PRFTINID is null since identifiers were not assigned to fetuses in pregnancies that ended before 20 weeks. |
| Row 3: | Shows the forceps-assisted delivery for the fetus identified with --FTINID="1". The classification of the forceps delivery as "LOW" was represented in the NSV PRBCLOC. |
pr.xpt
| Row | STUDYID | DOMAIN | USUBJID | PRSEQ | PRLNKID | PRTRT | PRCAT | PRSTDTC | PRFTINID | PRURGNCY | PRBCLOC | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC-123 | PR | 103 | 1 | PREG1 | Cesarean delivery | PREGNANCY-RELATED PROCEDURES | 2017-10-25 | 2 | EMERGENCY | ||
| 2 | ABC-123 | PR | 104 | 1 | PREG1 | Surgical ectopic pregnancy termination | PREGNANCY-RELATED PROCEDURES | 2017-05-15 | ||||
| 3 | ABC-123 | PR | 105 | 1 | PREG1 | Forceps-assisted delivery | PREGNANCY-RELATED PROCEDURES | 2017-06-01 | 1 | LOW |
PR NSV Metadata
| Variable | Label | Type | Role | Codelist | Origin |
|---|---|---|---|---|---|
| PRFTINID | Fetus/Infant Identifier | integer | Non-Standard Identifier | CRF | |
| PRURGNCY | Procedure Urgency Status Type | text | Non-Standard Record Qualifier | (PRURGNCY) | CRF |
| PRBCLOC | Birth Canal Location | text | Non-Standard Record Qualifier | CRF |
Pregnancies, pregnancy outcomes, and ends of pregnancies were represented in CE. Ends of pregnancies that were also procedures or medication administrations were also represented in the PR or CM domains. The RELREC links pregnancy-related records using --LNKID records at the maternal level. Linking procedures to a particular fetus also requires the use of --FTINID, but RELREC only allows 1 linking variable.
relrec.xpt
| Row | STUDYID | RDOMAIN | USUBJID | IDVAR | IDVARVAL | RELTYPE | RELID |
|---|---|---|---|---|---|---|---|
| 1 | ABC-123 | CE | CELNKID | MANY | 1 | ||
| 1 | ABC-123 | PR | PRLNKID | MANY | 1 | ||
| 1 | ABC-123 | CM | CMLNKID | ONE | 1 |
In a study in which mothers and infants were study subjects, pregnancy-related data would be represented as in the case when only the mother is a study subject. Note that since the information is represented in records for the mother, this information is associated with infants only indirectly. When a pregnancy includes only 1 fetus, the association between maternal and infant data would be adequately represented by the mother-infant relationships in RELSUB. For multiple pregnancies, the relationship between a fetal identifier in a maternal record and a USUBJID in an infant record (CEFTINID in the example above) cannot currently be represented in the SDTM. This relationship would be explained in a comment to USUBJID and/or SUBJID in the Define-XML document and/or in the Clinical Study Data Reviewers Guide (cSDRG).
In the studies on which the TAUG-HIV was based, only live-born infants were considered study subjects and prenatal data were treated as data about the mother. Other studies might take a different approach and treat fetuses as study subjects.
Example
This is an example of a study in which infants were study subjects, and any data about their mothers was represented in AP domains. In this example, pregnancy-related events were collected as in Example 1. However, because data on screen failures were not submitted, only data for mothers with live-born infants were submitted, and the resulting data were represented in APCE. The sponsor chose to base the mother's APID value on her infant's USUBJID, but with an appended "M". In the case of multiple infants, one infant's USUBJID was chosen for use in constructing the mother's APID. Note:
- This example shows data for twins, both of whom were born alive; in the multiple pregnancy in Example 1, only 1 twin survived. The current example also includes an example of a single birth.
- Although this example used a non-standard qualifier ("CEFTINID"), a sponsor could use a standard variable, such as CESPID, for this purpose. This example did not use CESPID in order to avoid creating the impression that a --SPID variable (which is sponsor-defined), should be used for the particular purpose illustrated in this example.
- Because maternal data was collected for only the pregnancy involving the infant study subject, there was no need to use RELREC to link records for a particular pregnancy.
| Row 1: | Shows a mother's pregnancy with 2 gestations, associated with 2 study subjects (101 and 102); RSUBJID is populated with "MULTIPLE". When an associated person has relationships to multiple subjects, a single value of "SREL" is usually inadequate; the SDTMIG-AP directs that SREL should be "MULTIPLE". In this case, the mother's relationship to both infants was the same, but the sponsor followed the rule in the SDTMIG-AP and populated RSUBJID with "MULTIPLE". The identifiers of the multiple subjects and the mother's relationship to each of them are in the APRELSUB dataset. |
|---|---|
| Rows 2-3: | Show data about the live birth and delivery of 1 twin. Because these records are associated with only one study subject, the sponsor populated SREL with the "MOTHER, BIOLOGICAL" and USUBJID with "101". |
| Rows 4-5: | Show data about the live birth of the other twin. This infant's cesarean delivery was also represented in the APPR domain. |
| Rows 6-8: | Show data about a singleton pregnancy. All data are associated with the subject with USUBJID = "103". This infant's forceps-assisted delivery was also represented in the APPR domain. |
apce.xpt
| Row | STUDYID | DOMAIN | APID | CESEQ | RSUBJID | SREL | CETERM | CECAT | CESCAT | CESTDTC | CEENDTC | CEFETNUM | CEFTINID | CESETTNG | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | XYZ-456 | CE | 101M | 1 | MULTIPLE | MULTIPLE | Pregnancy | PREGNANCY-RELATED EVENTS | 2017-01-26 | 2017-10-25 | 2 | ||||
| 2 | XYZ-456 | CE | 101M | 2 | 101 | MOTHER, BIOLOGICAL | Live birth | PREGNANCY-RELATED EVENTS | PREGNANCY OUTCOME | 2017-10-25 | 1 | VEHICLE | |||
| 3 | XYZ-456 | CE | 101M | 3 | 101 | MOTHER, BIOLOGICAL | Unassisted vaginal delivery | PREGNANCY-RELATED EVENTS | 2017-10-25 | 1 | |||||
| 4 | XYZ-456 | CE | 101M | 4 | 102 | MOTHER, BIOLOGICAL | Live birth | PREGNANCY-RELATED EVENTS | PREGNANCY OUTCOME | 2017-10-25 | 2 | HOSPITAL | |||
| 5 | XYZ-456 | CE | 101M | 5 | 102 | MOTHER, BIOLOGICAL | Cesarean delivery | PREGNANCY-RELATED EVENTS | 2017-10-25 | 2 | |||||
| 6 | XYZ-456 | CE | 103M | 1 | 103 | MOTHER, BIOLOGICAL | Pregnancy | PREGNANCY-RELATED EVENTS | 2016-09-15 | 2017-06-01 | 1 | ||||
| 7 | XYZ-456 | CE | 103M | 2 | 103 | MOTHER, BIOLOGICAL | Live birth | PREGNANCY-RELATED EVENTS | PREGNANCY OUTCOME | 2017-06-01 | 1 | HOSPITAL | |||
| 8 | XYZ-456 | CE | 103M | 3 | 103 | MOTHER, BIOLOGICAL | Forceps-assisted delivery | PREGNANCY-RELATED EVENTS | 2017-06-01 | 1 |
APCE NSV Metadata
| Variable | Label | Type | Role | Origin |
|---|---|---|---|---|
| CEFETNUM | Number of Fetuses/Infants in Pregnancy | integer | Non-Standard Record Qualifier | CRF |
| CEFTINID | Fetus/Infant Identifier | integer | Non-Standard Identifier | CRF |
| CESETTNG | Event Setting | text | Non-Standard Record Qualifier | CRF |
Cesarean and assisted deliveries were represented in the PR domain.
| Row 1: | Shows that for the infant identified with --FTINID = "2", the pregnancy ended in Cesarean delivery. The urgency of this procedure was represented with the non-standard variable PRURGNCY. |
|---|---|
| Row 2: | Shows the forceps-assisted delivery for the fetus identified with --FTINID = "1". The classification of the forceps delivery as "LOW" was represented in the NSV PRBCLOC. |
appr.xpt
| Row | STUDYID | DOMAIN | APID | PRSEQ | RSUBJID | SREL | PRTRT | PRCAT | PRSCAT | PRPRESP | PROCCUR | PRINDC | PRSTDTC | PRENDTC | PRSTDY | PRENDY | PRFTINID | PRURGNCY | PRBCLOC | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC-123 | PR | 101M | 1 | 102 | MOTHER, BIOLOGICAL | Cesarean delivery | PREGNANCY-RELATED PROCEDURES | 2017-10-25 | 2 | EMERGENCY | |||||||||
| 2 | ABC-123 | PR | 103M | 1 | 103 | MOTHER, BIOLOGICAL | Forceps-assisted delivery | PREGNANCY-RELATED PROCEDURES | 2017-06-01 | 1 | LOW |
APPR NSV Metadata
| Variable | Label | Type | Role | Codelist | Origin |
|---|---|---|---|---|---|
| PRFTINID | Fetus/Infant Identifier | integer | Non-Standard Identifier | CRF | |
| PRURGNCY | Procedure Urgency Status Type | text | Non-Standard Record Qualifier | (PRURGNCY) | CRF |
| PRBCLOC | Birth Canal Location | text | Non-Standard Record Qualifier | CRF |
5.4 Gestational Age
Estimations of gestational age may be based on examinations performed on a pregnant woman or on a newborn infant. Note that gestational age can be estimated at any time during pregnancy, although after birth "gestational age" is commonly used to refer to the gestational age at birth. Example 1 shows the representation of estimated gestational age, and Example 2 the representation of various kinds of data from which gestational age may be estimated.
Example
In this example, only infants were study subjects. However, with the possible exception of USUBJID values, the example would be unchanged for a study in which mothers were study subjects. If these data were collected for an infant who was an associated person, they would be represented in the APSC domain and the dataset would include APID, RSUBJID, and SREL, rather than USUBID.
Although there is a test for gestational age in the CDISC Controlled Terminology for Reproductive Findings, gestational age is an attribute of the fetus or infant, and is not a finding about their reproductive system; in this example, gestational age is represented in the Subject Characteristics (SC) domain. The structure of the SC domain is currently 1 record per characteristic (test) per subject, but it is expected that the structure will allow multiple records per test in the future. This example shows multiple estimates of gestational age for the same subject. In this example, estimated gestational age is shown as a finding about the mother, rather than the infant. In cases of a multiple-gestation pregnancies, separate estimates for different fetuses are distinguished using an identifier such as the non-standard identifier SCFTIND used in this example. Not all of the values of METHOD shown in this example are currently in the METHOD codelist.
Gestational age is often expressed (and sometimes collected) in weeks and days. SDTM does not support the recording of an individual finding result with mixed units (e.g., "20 weeks and 5 days"), so the gestational age would be converted to days for representation in SDTM.
| Rows 1-2: | Prenatal estimates of gestational age were obtained by 2 different methods at Visits 10 and 11. |
|---|---|
| Row 3: | Gestational age was estimated shortly after birth by physical examination of the infant. Birth did not occur during a regularly scheduled visit. Because it occurred between scheduled Visits 13 and 14, the sponsor assigned a visit number of 13.1 to data collected at that time. |
sc.xpt
| Row | STUDYID | DOMAIN | USUBJID | SCSEQ | SCTESTCD | SCTEST | SCCAT | SCORRES | SCORRESU | SCSTRESC | SCSTRESN | SCSTRESU | SCMETHOD | VISITNUM | SCDTC | SCDY | SCFTINID | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC-123 | SC | 101 | 1 | EGESTAGE | Estimated Gestational Age | PREGNANCY-RELATED FINDINGS | 100 | DAYS | 100 | 100 | DAYS | MENSTRUAL HISTORY | 10 | 2017-03-02 | 196 | 1 | |
| 2 | ABC-123 | SC | 101 | 2 | EGESTAGE | Estimated Gestational Age | PREGNANCY-RELATED FINDINGS | 135 | DAYS | 135 | 135 | DAYS | ULTRASOUND | 11 | 20-17-04-01 | 226 | 1 | |
| 3 | ABC-123 | SC | 101 | 3 | EGESTAGE | Estimated Gestational Age | PREGNANCY-RELATED FINDINGS | 265 | DAYS | 265 | 265 | DAYS | BALLARD | 13.1 | 20-17-06-10 | 297 | 1 |
SC NSV Metadata
| Variable | Label | Type | Role | Origin |
|---|---|---|---|---|
| SCFTINID | Fetus/Infant Identifier | text | Non-Standard Identifier | CRF |
If data to support an estimate of gestational age are collected, the domain in which those data are represented depends on the nature of the supporting data. If study data for estimated gestational age did have supporting data, the relationship between the estimated gestational age record in SC and the supporting data in another domain would be represented in RELREC.
The Ballard score is an assessment of physical characteristics of a newborn from which gestational age may be estimated. This is a clinical classification that would be represented in the Disease Response and Clinical Classification (RS) domain. Development of a supplement for this scale has been requested. See Section 7.6, Questionnaires, Ratings, and Scales, for further information about QRS supplements.
Gestational age may be estimated from the date of the last menstrual period, which is the start date of the event "Menstrual period". In a study that enrolled pregnant women, the most recent menstrual period would be represented in MH, using a form similar to that shown in Section 4.1, Baseline Menstrual History. In a study where the last menstrual period occurred during the study, it would be represented in the Clinical Events (CE) domain.
Example
In this example, a pregnant woman was a study subject. Whether infants were study subjects would not affect this example.
In this example, the date of the start of the last menstrual period was collected if a woman became pregnant during the study. The non-standard variable CECRNORD="MOST RECENT" represents the relationship of the particular menstrual period relative to the date of collection (CEDTC). The collected start date is represented in CESTDTC.
ce.xpt
| Row | STUDYID | DOMAIN | USUBJID | CESEQ | CEGRPID | CETERM | CECAT | CEDTC | CESTDTC | CECRNORD | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC-123 | CE | 101 | 1 | PREG1 | Menstrual period | PREGNANCY-RELATED EVENTS | 2017-05-15 | 2017-03-01 | MOST RECENT | |
| 2 | ABC-123 | CE | 101 | 2 | PREG2 | Menstrual period | PREGNANCY-RELATED EVENTS | 2017-09-02 | 2017-07-23 | MOST RECENT |
CE NSV Metadata
| Variable | Label | Type | Role | Origin |
|---|---|---|---|---|
| CECRNORD | Chronological Order | text | Non-Standard Record Qualifier | CRF |
Fundal height, which is the distance from the top of the pubic bone to the top of the uterus assessed by palpation, can be used to estimate gestational age. Because this test is a measurement of the uterus, it is represented in the Reproductive System Findings (RP) domain.
Example
In this example, a pregnant woman was a study subject. Whether infants were study subjects would not affect this example.
The values for RPTEST and RPTESTCD shown in this example are not in current published controlled terminology. The value of RPCAT reflects the name of the CRF module in which this measurement was collected.
rp.xpt
| Row | STUDYID | DOMAIN | USUBJID | RPSEQ | RPGRPID | RPTESTCD | RPTEST | RPCAT | RPORRES | RPORRESU | RPSTRESC | RPSTRESN | RPSTRESU | VISITNUM | RPDTC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC-123 | RP | 103 | 1 | PREG1 | FUNDHT | Fundal Height | PREGNANCY-RELATED EVENTS | 28 | cm | 28 | 28 | cm | 4 | 2016-07-13 |
Gestational age can also be estimated from ultrasound measurements of a fetus in utero. Body measurements such as head circumference and bone lengths are represented in the Vital Signs (VS) domain.
Example
In this example, a pregnant woman was a study subject. Whether infants were study subjects would not affect this example. This example shows only 1 of the measurements that may be used to estimate gestational age. Fetal ultrasound measurements were treated as findings about the mother, who undergoes the procedure; results are generally entered in the mother's charts. In the case of multiple births, fetal measurements are distinguished from each other, but measurements made on different occasions may not be associated with the same fetus. That is, measurements at different times might be recorded as for Fetus 1 and Fetus 2, but Fetus 1 at the 2 occasions may or may not be the same fetus. Given this situation, it did not seem appropriate to assign an APID (or a USUBJID, depending on the study situation) to fetal measurements. This might be approached differently in a study in which an effort is made to assign invariant identifiers to fetuses. In this example, the non-standard identifier VSFTINID was used to distinguish between infants in a multiple-gestation pregnancy. The VSSPID variable was considered for this purpose but was not used, because --SPID is a sponsor-defined variable for which a sponsor may have other uses.
The test terminology shown here, which includes "fetal" to describe the measurements, was not in CDISC Controlled Terminology at the time of publication of this user guide, but the terms are similar to existing SEND controlled terms such as "Fetal Body Weight". In cases of multiple-gestation pregnancies, separate estimates for different fetuses are distinguished using the non-standard identifier VSFTINID.
vs.xpt
| Row | STUDYID | DOMAIN | USUBJID | VSSEQ | VSGRPID | VSTESTCD | VSTEST | VSCAT | VSORRES | VSORRESU | VSSTRESC | VSSTRESN | VSSTRESU | VISITNUM | VSDTC | VSFTINID | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC-123 | VS | 103 | 1 | PREG1 | FTHDCIRC | Fetal Head Circumference | PREGNANCY-RELATED EVENTS | 25 | cm | 25 | 25 | cm | 3 | 2016-04-16 | 1 | |
| 2 | ABC-123 | VS | 103 | 2 | PREG1 | FTHDCIRC | Fetal Head Circumference | PREGNANCY-RELATED EVENTS | 28 | cm | 28 | 28 | cm | 3 | 2016-04-16 | 2 |
VS NSV Metadata
| Variable | Label | Type | Role | Origin |
|---|---|---|---|---|
| VSFTINID | Fetus/Infant Identifier | integer | Non-Standard Identifier | CRF |
5.5 Infant Demographics Data
In studies in which the infant is considered an associated person, demographics data will be limited. Many of the date variables in the Demographics (DM) domain are dates that refer to study participation, which do not apply to an associated person and would not be included in an APDM dataset.
Example
In this example, mothers were study subjects, and any data about infants were represented in associated persons (AP) domains.
In this example, only live-born infants were considered to be associated persons; data about fetuses who were not born live was considered maternal data. Birth date and sex were the only demographic data collected about infants. Other demographic data were not included because they are neither expected nor required in the APDM domain.
Controlled terminology values for the relationship of an associated person to a study subject include the gender-neutral term "CHILD, BIOLOGICAL" as well as the terms "DAUGHTER, BIOLOGICAL" and "SON, BIOLOGICAL". In this example, the gender-neutral term was used for all infants, but it would also be possible and valid to use the gender-specific terms.
apdm.xpt
| Row | STUDYID | DOMAIN | APID | RSUBJID | SREL | BRTHDTC | SEX |
|---|---|---|---|---|---|---|---|
| 1 | ABC-123 | APDM | 103-2 | 103 | CHILD, BIOLOGICAL | 2017-10-25 | M |
| 2 | ABC-123 | APDM | 105-1 | 105 | CHILD, BIOLOGICAL | 2017-06-01 | F |
Demographics data include multiple dates related to study participation. In studies that recruit pregnant women and in which mothers and infants are study subjects, the meaning of these dates for an infant subject should be defined in the protocol:
- The date of informed consent (i.e., informed assent for the infant), RFICDTC, will be before birth.
- The dates of start and end of study treatment, RFXSTDTC and RFXENDTC, would depend on whether infants receive study treatment directly and whether indirect exposure in utero or via breast milk is included.
- The study reference start date, RFSTDTC, should have the same definition for all study subjects, and have a meaning that will make --DY variables that reference this date useful. This choice may be difficult in studies that include both mothers and infants as study subjects.
Study participation dates in infant demographic data for studies in which only the infant is a study subject may be somewhat easier to define.
For instance, in the infant PK study example in Section 5.1, Studies Involving Mother-Infant Data, the study reference start date might be chosen as the date of birth or the date of start of direct study treatment.
5.6 Assessments of Infants at Birth
The examples in this section assume that the infant is a study subject. Data collected for an infant who was an associated person would be represented in the corresponding AP domain and the dataset would include APID, RSUBJID, and SREL, rather than USUBID.
Apgar scores and Ballard Maturational Assessments may be performed shortly after birth. Examples of data from these scales are not included here, as examples would be developed by the Questionnaires, Ratings, and Scales (QRS) Team. See Section 7.6, Questionnaires, Ratings, and Scales for additional information about these and other scales relevant to HIV studies.
Congenital anomalies would be recorded as adverse events for the infant.
Infant weight may be compared to a growth chart to determine whether an infant is small for gestational age (generally defined as having a weight less than the 10th percentile for gestational age).
Example
In this example, infants were study subjects. The USUBJID values in the example suggest that mothers were also study subjects, but with the possible exception of USUBJID values, the example would be unchanged for a study in which mothers were associated persons.
In this example, infants were weighed at birth, and their weight was compared to a growth chart to determine whether they were small for gestational age. The date of collection is the infant's date of birth, which can be obtained from the Demographics (DM) record for the infant. The infant in this example had a gestational age of 280 days (40 weeks), as determined by the Ballard method of examination, and was considered small for gestational age..
vs.xpt
| Row | STUDYID | DOMAIN | USUBJID | VSSEQ | VSTESTCD | VSTEST | VSORRES | VSORRESU | VSSTRESC | VSSTRESN | VSSTRESU | VSDTC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC-2 | VS | 105A | 1 | WEIGHT | Weight | 2050 | g | 2050 | 2050 | g | 2015-02-03 |
| Row 1: | Shows gestational age for an infant using the Ballard examination, represented by SCMETHOD="BALLARD". |
|---|---|
| Row 2: | Shows that an infant was determined to be small for gestational age. SCMETHOD="FENTON 2013" was used to indicate the growth chart to which the infant's weight and gestational age were compared. |
sc.xpt
| Row | STUDYID | DOMAIN | USUBJID | SCSEQ | SCTESTCD | SCTEST | SCORRES | SCORRESU | SCSTRESC | SCSTRESN | SCSTRESU | SCMETHOD | SCDTC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC-2 | SC | 105A | 1 | EGESTAGE | Estimated Gestational Age | 280 | DAYS | 280 | 280 | DAYS | BALLARD | 2015-02-03 |
| 2 | ABC-2 | SC | 105A | 2 | SMGAIND | Small for Gestational Age Indicator | Y | Y | FENTON 2013 | 2015-02-03 |
Fontanelle closure may also be assessed at birth.
Example
In this example, both mothers and infants were study subjects. However, with the possible exception of USUBJID values, the example would be unchanged for a study in which mothers were not study subjects.
The infant had a postnatal examination to assess the closure of the anterior and posterior fontanelle. The MKTEST, MKTESTCD, and MKLOC values shown in this example are not yet included in published controlled terminology at the time of publication of this user guide.
mk.xpt
| Row | STUDYID | DOMAIN | USUBJID | MKSEQ | MKTESTCD | MKTEST | MKCAT | MKORRES | MKSTRESC | MKLOC | MKDTC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC-2 | MK | 101A | 1 | FONTCLOS | Fontanelle Closure Indicator | POSTNATAL EXAM | N | N | ANTERIOR FONTANELLE | 2015-01-24T13:24 |
| 2 | ABC-2 | MK | 101A | 2 | FONTCLOS | Fontanelle Closure Indicator | POSTNATAL EXAM | N | N | POSTERIOR FONTANELLE | 2015-01-24T13:24 |
5.7 Infant Feeding
In studies where the infant is a study subject, data may be collected about infant feeding. Data about breastfeeding may be used in combination with maternal medication data to assess infant exposure to medications in breast milk.
Example
This is an example of a study in which infants were study subjects. The USUBJID values suggest that mothers were also study subjects; with the possible exception of USUBJID values, the example would be unchanged for a study in which mothers were associated persons. If these data were collected for an infant who was an associated person, they would be represented in the APML domain and the dataset would include APID, RSUBJID, and SREL, rather than USUBID.
In this study, data about infant feeding included:
- An indication of whether the infant was breastfed
- If the infant was not breastfed, the reason for not breastfeeding was collected.
- If the infant was breastfed, the start and end dates of feeding with breast milk were collected.
- An indication of whether the infant received formula
- If the infant received formula, the start and end dates of feeding with formula were collected.
- An indication of whether the infant received complementary food (i.e., food other than breast milk or formula)
- If the infant received complementary food, the start date of feeding with complementary food was collected.
Data are shown for 2 infants; 1 was breastfed and the other was not.
ml.xpt
| Row | STUDYID | DOMAIN | USUBJID | MLSEQ | MLTRT | MLPRESP | MLOCCUR | MLSTDTC | MLENDTC | MLREASOC | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC-2 | ML | 101A | 1 | BREAST MILK | Y | Y | 2016-03-18 | 2017-02-13 | ||
| 2 | ABC-2 | ML | 101A | 2 | FORMULA | Y | Y | 2016-08-25 | |||
| 3 | ABC-2 | ML | 101A | 3 | COMPLEMENTARY FOOD | Y | Y | 2017-01-25 | |||
| 4 | ABC-2 | ML | 102A | 1 | BREAST MILK | Y | N | MATERNAL ILLNESS | |||
| 5 | ABC-2 | ML | 102A | 2 | FORMULA | Y | Y | 2016-04-01 | |||
| 6 | ABC-2 | ML | 102A | 3 | COMPLEMENTARY FOOD | Y | Y | 2017-02-04 |
ML NSV Metadata
| Variable | Label | Type | Role | Origin |
|---|---|---|---|---|
| MLREASOC | Reason for Occurrence Value | text | Non-Standard Record Qualifier | CRF |
5.8 Routine Data for Mother-Infant Pairs
The SDTMIG provides advice on the collection of data such as vital signs, adverse events, and medications, and the SDTMIG-AP provides information on how to represent these kinds of data for associated persons. The following are a few points to consider in a study that involves mother-infant pairs.
Vital Signs
There is no specific test for "birth weight". Birth weight would be represented in a Vital Signs (VS) or APVS domain in a record with VSDTC equal to the birth date.
Medical Conditions
For study subjects, medical conditions are represented in Medical History (MH), Adverse Events (AE), or Clinical Events (CE), depending on when they occurred (MH for events that started before the study) and on whether the event is reportable as an adverse event in the particular study (the distinction between AE and CE). For associated persons, the distinctions are less clear. APMH tends to be used for medical conditions before the study subject started the study. For mothers who are associated persons for an infant study subject, however, the domain in which to represent events that occurred after the pregnant woman was recruited might be represented in CE or AE, even though the infant may not be considered to start the study until after birth.
Example
In this example, the mother is an associated person. If the mother were a study subject, the data would be represented in the MH domain and the dataset would include USUBID, rather than APID, RSUBJID, and SREL. The sponsor chose to represent these maternal events as medical history, because they occurred before the subject was born.
| Rows 1-2: | Show records for conditions which were prespecified in a CRF module for pregnancy complications. MHCAT="PREGNANCY COMPLICATIONS" to indicate the source module. |
|---|---|
| Rows 3-4: | Show conditions that were not prespecified. |
apmh.xpt
| Row | STUDYID | DOMAIN | APID | MHSEQ | RSUBJID | SREL | MHTERM | MHDECOD | MHCAT | MHPRESP | MHOCCUR | MHSTDTC | MHENDTC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC-3 | APMH | 107M | 1 | 107 | MOTHER, BIOLOGICAL | Preeclampsia | Preeclampsia | PREGNANCY COMPLICATIONS | Y | Y | 2015-09-03 | 2015-11-25 |
| 2 | ABC-3 | APMH | 107M | 2 | 107 | MOTHER, BIOLOGICAL | Premature labor | Premature labor | PREGNANCY COMPLICATIONS | Y | N | ||
| 3 | ABC-3 | APMH | 107M | 3 | 107 | MOTHER, BIOLOGICAL | Fever | Fever | 2015-10-05 | 2015-10-09 | |||
| 4 | ABC-3 | APMH | 107M | 4 | 107 | MOTHER, BIOLOGICAL | Cough | Cough | 2016-01-08 | 2016-10-08 |
Medications
Maternal medications during pregnancy and lactation are likely to be of interest when the infant is a study subject. Which medications were taken during pregnancy and lactation may be ascertained by comparing intervention record dates with the date of birth and with dates of breastfeeding. When the mother is an associated person rather than a study subject, limited concomitant medication data might be collected, and the data might be collected in separate modules for medications taken during pregnancy and those taken during lactation. If so, then the collection module of origin could be represented in a value of the CMCAT or CMSCAT variable.
Example
In this example, the mother was an associated person. If she were an study subject, the data would be represented in the CM domain and the dataset would include USUBID, rather than APID, RSUBJID, and SREL. Data about her medications during breastfeeding were collected in a separate CRF module, and the name of module was used to populate CMCAT. This example shows modeling of a combination therapy with each drug and dosage listed separately. Either CMENRF or CMENRTPT can be used to represent data coming from the CMONGO variable. In this case, CMENRF was populated with PRIOR because the concomitant medications were not ongoing.
The Medication During Breastfeeding CRF was used in this infant PK study to collect information on medications taken by a mother.
Because the mother is not the actual subject, the Associated Persons SDTMIG domain APCM would be used. Within APCM, variable names use the Concomitant/Prior Medications (CM) domain prefix. Consult the SDTMIG and CDASHIG for CM variable-level metadata.
Where variable metadata do not already exist in the CDASHIG, the CDASH Implementation Guide provides guidance on developing variables based upon the CDASH and SDTM models and the SDTMIG.
The following are required SDTMIG variables for the APCM domain:
- APID (Associated Persons Identifier)
- SREL (Subject, Device or Study Relationship), prepopulated in this example with the value of "MOTHER, BIOLOGICAL" from the RELSUB codelist
Breastfeeding Medication Log
| Indicate if any concomitant medications were taken. If Yes, include the appropriate details where indicated on the CRF. | Any Concomitant Medication(s)
CMYN Not submitted |
<From NY codelist> |
|---|---|---|
| Record only 1 medication per line. Provide the full trade or proprietary name of the medication; otherwise, the generic name may be recorded. |
CMTRT | _________________ |
| Record the concomitant medication subcategory. | What is the subcategory for the concomitant medication?
CMSCAT |
|
| Record the dose of concomitant medication taken per administration (e.g., 200). |
CMDOSE | _________________ |
| Record the dose unit of the dose of concomitant medication taken (e.g., mg). |
CMDOSU | _________________
<From UNIT codelist> |
| Record how often the concomitant medication was taken (e.g., BID, PRN). |
CMDOSFRQ | _________________
<From FREQ codelist> |
| Provide the route of administration for the concomitant medication. |
CMROUTE | _________________
<From ROUTE codelist> |
| Record the date the concomitant medication was first taken, using this format: DD-MON-YYYY. If the subject has been taking the concomitant medication for a considerable amount of time prior to the start of the study, it is acceptable to have an incomplete date. Any concomitant medication taken during the study is expected to have a complete start date. Any prior concomitant medication that is exclusionary should have both a start and end date. |
CMSTDAT CMSTDTC | _________________ |
| Record the concomitant medication as ongoing or not, to indicate whether the subject has stopped taking the concomitant medication at the time of data collection. If the concomitant medication is ongoing, the end date should be left blank. | Was the concomitant medication ongoing?
CMONGO CMENRF/CMENRTPT |
<From NY codelist> |
| Record the date the concomitant medication was stopped, using this format: DD-MON-YYYY. If the subject has not stopped taking the concomitant medication, leave this field blank. |
CMENDAT CMENDTC | _________________ |
CRF Metadata
apcm.xpt
| Row | STUDYID | DOMAIN | APID | CMSEQ | RSUBJID | SREL | CMTRT | CMCAT | CMSCAT | CMDOSE | CMDOSU | CMDOSFRQ | CMROUTE | CMDTC | CMSTDTC | CMENDTC | CMENRF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC | APCM | 107 | 1 | 107A | MOTHER, BIOLOGICAL | Efavirenz | MEDICATION DURING BREASTFEEDING | ANTIRETROVIRAL | 600 | mg | QD | ORAL | 2015-11-26 | 2015-09-03 | 2015-11-25 | PRIOR |
| 2 | ABC | APCM | 107 | 2 | 107A | MOTHER, BIOLOGICAL | Emtricitabine | MEDICATION DURING BREASTFEEDING | ANTIRETROVIRAL | 200 | mg | QD | ORAL | 2015-11-26 | 2015-09-03 | 2015-11-25 | PRIOR |
| 3 | ABC | APCM | 107 | 3 | 107A | MOTHER, BIOLOGICAL | Tenofovir disoproxil fumarate | MEDICATION DURING BREASTFEEDING | ANTIRETROVIRAL | 300 | mg | QD | ORAL | 2015-11-26 | 2015-09-03 | 2015-11-25 | PRIOR |
If the infant is a study subject, exposure to maternal medications may be represented in interventions domains. For these medications, however, the dose taken by the infant would not be known, so some dose-related domain variables would not be populated..
Example
In this example, mothers and infants were study subjects. However, with the possible exception of USUBJID values, the example would be unchanged for a study in which mothers were not study subjects. If these data were collected for an infant who was an associated person, they would be represented in the APML domain and the dataset would include APID, RSUBJID, and SREL, rather than USUBID.
This example shows infant exposure to study drug before birth and during breastfeeding. Because infant exposure data is derived from maternal data, only the EX dataset is shown. Since EXDOSE, EXDOSU, and EXDOSFRM are expected, they are included though not populated.
ex.xpt
| Row | STUDYID | DOMAIN | USUBJID | EXSEQ | EXTRT | EXDOSE | EXDOSU | EXDOSFRM | EXROUTE | EXSTDTC | EXENDTC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC-2 | EX | 101A | 1 | CURALL | TRANSPLACENTAL | 2016-03-25 | 2016-07-04 | |||
| 2 | ABC-2 | EX | 101A | 2 | CURALL | TRANSMAMMARY | 2016-07-04 | 2016-12-18 |
6 Interventions for the Prevention and Treatment of HIV
6.1 HIV Treatment with Oral Antiretroviral Therapy (ART)
The goal of ART in the treatment of HIV is to slow progression of the disease by curbing the proliferation of the virus. Successful control of HIV by ART can significantly prolong life (as compared to uncontrolled HIV infection), as well as reduce the chances of the infected person passing the virus to others. ART consists of a combination of antiretroviral drugs (ARVs). Therapy of a single ARV is not used; they are always given in combinations consisting of multiple ARVs. For instance, recommendations for the initial treatment of HIV infection call for a combination of 3 or more ARVs from 2 or more drug classes.[11]
Handling data on drug regimens involving combination therapies can be complex and calls for more specific data modeling strategies than those illustrated in the current SDTMIG. In order to develop a unified approach to handling these data across all therapeutic areas, a stand-alone supplemental focused topic guide on combination therapies will be developed to include examples from multiple disease areas. Users should refer to the HIV ART example(s) in that guide for further information when that topic guide is published.
6.2 Pre-exposure Prophylaxis (PrEP)
Pre-exposure prophylaxis (PrEP) is defined as receiving antiviral drug(s) prior to exposure to prevent infection. Subjects receive ARV therapy prior to exposure to HIV to reduce the risk of becoming infected. PrEP may be an option for subjects who have a high risk of becoming infected with HIV and it may be taken for varying lengths of time, depending on risk.
Section 6.2.1, Vaginal Ring, provides an example of a vaginal ring containing an antiviral drug as a form of PrEP. Oral PrEP examples are not provided in the TAUG-HIV as the representation of this treatment is well handled by both the CDASHIG and the SDTMIG.
6.2.1 Vaginal Ring
Vaginal rings are one form of PrEP and have been shown to be an effective tool for women to reduce the risk of HIV transmission from infected sexual partners. The silicone ring is inserted into the vagina and it continuously releases an antiviral drug over the period of a month.
Example
In a hypothetical prevention study, investigators assessed the safety and efficacy of a vaginal ring containing approximately 25 mg of Study Drug A to prevent HIV infection. The study was conducted over a 3-month period, and women were instructed to insert each ring for 28 days. If the ring was removed for any reason, they were asked to record the information about the removal, including the removal date, reason for the removal, whether or not the ring was washed, and whether or not they reinserted the ring. After the 28 days, they were asked to return to the study site so that the ring could be removed and replaced. At this time, subjects provided a blood sample in order to determine the pharmacokinetic concentration of Drug A. The amount of Drug A remaining in the ring was also assessed at this time.
The following example shows study data from one subject (ABC-001) as described above over a 3-month period. The Device Identifiers (DI) domain shows identifying information for the 3 different rings dispensed to Subject ABC-001, including device type, lot number, and batch number. Each ring has a unique identifier, which is represented in the variable SPDEVID. This variable is used in several dataset examples below to identify data associated with each ring.
di.xpt
| Row | STUDYID | DOMAIN | SPDEVID | DISEQ | DIPARMCD | DIPARM | DIVAL |
|---|---|---|---|---|---|---|---|
| 1 | ABC | DI | RING583 | 1 | DEVTYPE | Device Type | Intravaginal Ring |
| 2 | ABC | DI | RING583 | 2 | LOT | Lot Number | ABC123 |
| 3 | ABC | DI | RING583 | 3 | BATCH | Batch Number | 6789 |
| 4 | ABC | DI | RING584 | 1 | DEVTYPE | Device Type | Intravaginal Ring |
| 5 | ABC | DI | RING584 | 2 | LOT | Lot Number | ABC456 |
| 6 | ABC | DI | RING584 | 3 | BATCH | Batch Number | 6710 |
| 7 | ABC | DI | RING585 | 1 | DEVTYPE | Device Type | Intravaginal Ring |
| 8 | ABC | DI | RING585 | 2 | LOT | Lot Number | ABC789 |
| 9 | ABC | DI | RING585 | 3 | BATCH | Batch Number | 6705 |
The Device Properties (DO) domain was used to represent information about the amount of study drug in each of the three rings before insertion. The amount of starting drug in a ring may vary slightly by batch. As shown in the preceding DI example, each ring comes from a different batch.
do.xpt
| Row | STUDYID | DOMAIN | SPDEVID | DOSEQ | DOTESTCD | DOTEST | DOORRES | DOORRESU | DOSTRESC | DOSTRESN | DOSTRESU |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC | DO | RING583 | 1 | DRUGAAMT | Drug A Amount | 24.978 | mg | 24.978 | 24.978 | mg |
| 2 | ABC | DO | RING584 | 1 | DRUGAAMT | Drug A Amount | 24.854 | mg | 24.854 | 24.854 | mg |
| 3 | ABC | DO | RING585 | 1 | DRUGAAMT | Drug A Amount | 24.975 | mg | 24.975 | 24.975 | mg |
The Device Exposure (DX) domain was used to show each time the was ring was inserted and removed. Two non-standard variables (--INSROL and --RMVROL) indicate the role of person who inserted or removed the ring, respectively. If the ring fell out on its own, the non-standard variable "--RMVROL" was left null. These 2 non-standard variables were populated using the CDISC "Evaluator" codelist. The reason for ring removal was represented by the standard variable --RSDISC (Reason for Treatment Discontinuation). If a ring is removed and then reinserted, it may be important to collect whether or not the ring was rinsed before reinsertion, as this may alter the amount of drug that is in the ring. The non-standard variable --RNSIND was used to represent this concept.
dx.xpt
| Row | STUDYID | DOMAIN | USUBJID | SPDEVID | DXSEQ | DXTRT | DXRSDISC | DXSTDTC | DXENDTC | DXINSROL | DXRMVROL | DXRNSIND | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC | DX | ABC-001 | RING583 | 1 | Intravaginal Ring A | RING FELL OUT | 2016-02-01 | 2016-02-22 | HEALTH CARE PROFESSIONAL | N | ||
| 2 | ABC | DX | ABC-001 | RING583 | 2 | Intravaginal Ring A | RING DEPLETED | 2016-02-26 | 2016-02-28 | STUDY SUBJECT | HEALTH CARE PROFESSIONAL | Y | |
| 3 | ABC | DX | ABC-001 | RING584 | 3 | Intravaginal Ring A | RING DEPLETED | 2016-02-28 | 2016-03-26 | HEALTH CARE PROFESSIONAL | HEALTH CARE PROFESSIONAL | N | |
| 4 | ABC | DX | ABC-001 | RING585 | 4 | Intravaginal Ring A | STUDY SUBJECT DECLINED | 2016-03-26 | 2016-04-10 | HEALTH CARE PROFESSIONAL | STUDY SUBJECT | N |
DX NSV Metadata
| Variable | Label | Type | Role | Origin |
|---|---|---|---|---|
| DXINSROL | Role of Person Who Inserted the Device | text | Non-Standard Record Qualifier | CRF |
| DXRMVROL | Role of Person Who Removed the Device | text | Non-Standard Record Qualifier | CRF |
| DXRNSIND | Device Rinsed Indicator | text | Non-Standard Record Qualifier | CRF |
After the final removal of each ring, the concentration of drug found in the subject's plasma as well as the amount remaining in the ring was assessed. Ideally, the subject and the ring are assessed 28 days after the ring is initially inserted; thus the --TPT and --ELTM variables for each measurement were populated with the planned time of "28 DAYS AFTER RING INSERTION" and P28D, respectively. The variables --TPTREF and --RFTDTC, which are used to represent the ring insertion date, are also populated.
The results from the subject were represented in the Pharmacokinetics Concentrations (PC) domain, whereas the results from the ring were represented in the Device in Use (DU) domain, as shown in the example datasets below.
pc.xpt
| Row | STUDYID | DOMAIN | USUBJID | SPDEVID | PCSEQ | PCTESTCD | PCTEST | PCORRES | PCORRESU | PCSTRESC | PCSTRESN | PCSTRESU | PCSPEC | PCDTC | VISITNUM | VISIT | PCTPT | PCTPTNUM | PCELTM | PCTPTREF | PCRFTDTC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC | PC | ABC-101 | RING583 | 1 | DRUGA | Drug A | 200 | pg/mL | 200 | 200 | pg/mL | PLASMA | 2016-02-28 | 2 | VISIT 2 | 28 DAYS AFTER RING INSERTION | 1 | P28D | RING 1 INSERTION | 2016-02-01 |
| 2 | ABC | PC | ABC-101 | RING584 | 2 | DRUGA | Drug A | 264 | pg/mL | 264 | 264 | pg/mL | PLASMA | 2016-03-26 | 3 | VISIT 3 | 28 DAYS AFTER RING INSERTION | 1 | P28D | RING 2 INSERTION | 2016-02-28 |
| 3 | ABC | PC | ABC-101 | RING585 | 3 | DRUGA | Drug A | 50 | pg/mL | 50 | 50 | pg/mL | PLASMA | 2016-04-10 | 4 | VISIT 4 | 28 DAYS AFTER RING INSERTION | 1 | P28D | RING 3 INSERTION | 2016-03-26 |
du.xpt
| Row | STUDYID | DOMAIN | SPDEVID | DUSEQ | DUTESTCD | DUTEST | DUORRES | DUORRESU | DUSTRESC | DUSTRESN | DUSTRESU | DUDTC | VISITNUM | VISIT | DUTPT | DUTPTNUM | DUELTM | DUTPTREF | DURFTDTC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC | DU | RING583 | 1 | DRUGAAMT | Drug A Amount | 6.345 | mg | 6.345 | 6.345 | mg | 2016-02-28 | 2 | VISIT 2 | 28 DAYS AFTER RING INSERTION | 1 | P28D | RING 1 INSERTION | 2016-02-01 |
| 2 | ABC | DU | RING584 | 1 | DRUGAAMT | Drug A Amount | 3.557 | mg | 3.557 | 3.557 | mg | 2016-03-26 | 3 | VISIT 3 | 28 DAYS AFTER RING INSERTION | 1 | P28D | RING 2 INSERTION | 2016-02-28 |
| 3 | ABC | DU | RING585 | 1 | DRUGAAMT | Drug A Amount | 20.083 | mg | 20.083 | 20.083 | mg | 2016-04-10 | 4 | VISIT 4 | 28 DAYS AFTER RING INSERTION | 1 | P28D | RING 3 INSERTION | 2016-03-26 |
In this example, an EX dataset was derived by calculating the difference between the records in DO (e.g., the drug load levels) and DU (e.g., the ring residual drug level) data, and thus the participant's exposure to Drug A. Informative metadata regarding the derivation should be provided in the SDTM Define-XML file.
This example should not be taken as regulatory guidance regarding how a sponsor should derive an EX dataset.
ex.xpt
| Row | STUDYID | DOMAIN | USUBJID | SPDEVID | EXSEQ | EXTRT | EXDOSE | EXDOSU | EXDOSFRM | EXDOSFRQ | EXROUTE | EXSTDTC | EXENDTC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC | EX | ABC-101 | RING583 | 1 | Drug A | 18.633 | mg | RING | CONTINUOUS | INTRAVAGINAL | 2016-02-01 | 2016-02-28 |
| 2 | ABC | EX | ABC-101 | RING584 | 2 | Drug A | 21.297 | mg | RING | CONTINUOUS | INTRAVAGINAL | 2016-02-28 | 2016-03-26 |
| 3 | ABC | EX | ABC-101 | RING585 | 3 | Drug A | 4.892 | mg | RING | CONTINUOUS | INTRAVAGINAL | 2016-03-26 | 2016-04-10 |
6.3 Vaccines
As with any vaccine, the goal of a future vaccine against HIV will be to produce an immune response to the virus in order to prevent infection. As of the publication of this guide, no successful vaccine against HIV has been developed. There are numerous challenges to HIV vaccine development which are not discussed here, but research is ongoing. This section covers some of the topics relevant to those studies, including reactogenicity and vaccine-induced seroreactivity (VISR).
6.3.1 Reactogenicity
Vaccines are preparations containing antigenic substances capable of inducing a specific and active immunity against a disease or infection. Such immunity may then result in the reduced transmission of that disease or infection.
An important aspect of vaccine development is the assessment of the vaccine's reactogenicity. Reactogenicity refers to the property of a substance to produce an expected or common adverse reaction when introduced into the body. For the purpose of this guide, reactogenicity event refers to a specific expected or common reaction following vaccine administration. In vaccine studies, reactogenicity events are typically caused by an inflammatory response to the vaccine under study and may include reactions such as fever or redness at the site of administration. Reactogenicity describes immediate, short-term reactions to vaccines, not long-term sequelae.
For guidance on how to represent vaccine reactogenicity events for HIV, please refer to the Vaccines Therapeutic Area User Guide v1.1 (https://www.cdisc.org/standards/therapeutic-areas/vaccines).
6.3.2 Immune Response
Vaccines against HIV infection may be designed to raise antibodies against a number of specific epitopes on various HIV antigens. Measuring antibody titer would be one way to assess vaccine response in HIV vaccine trials.
Data about immune response to vaccines would be represented in the Immunogenicity Specimen Assessment (IS) domain.
For examples illustrating how to represent immune titers in response to vaccines, see the Virology Therapeutic Area User Guide v2.1 (https://www.cdisc.org/standards/therapeutic-areas/virology). Implementers should be aware that terminology specific to the titers they are performing may not yet be developed, and that new terminology can be requested using the New Term Request feature on the National Cancer Institute Enterprise Vocabulary Services website (https://www.cancer.gov/research/resources/terminology/cdisc).
For examples of anti-HIV antibody-specific terminology requested as part of this document, see the mb.xpt example within Section 6.3.3, Vaccine-induced Seroreactivity, Example 1.
6.3.3 Vaccine-induced Seroreactivity
Although no vaccine for HIV has been successful at the time of publication of this guide, multiple studies have been and continue to be conducted. In the post-vaccine world of HIV testing, an additional possible outcome to be considered is VISR. This outcome results from having been exposed to certain HIV antigens through a vaccine, as opposed to having been exposed to the actual virus. Given the stigma associated with HIV-positive status, it is important to distinguish between true positivity and seropositivity due to vaccine exposure.
The following concept map depicts a widely accepted algorithmic approach to determining HIV status, with considerations for interpreting results within the context of prior HIV vaccination.
Concept Map. Vaccine-induced Seroreactivity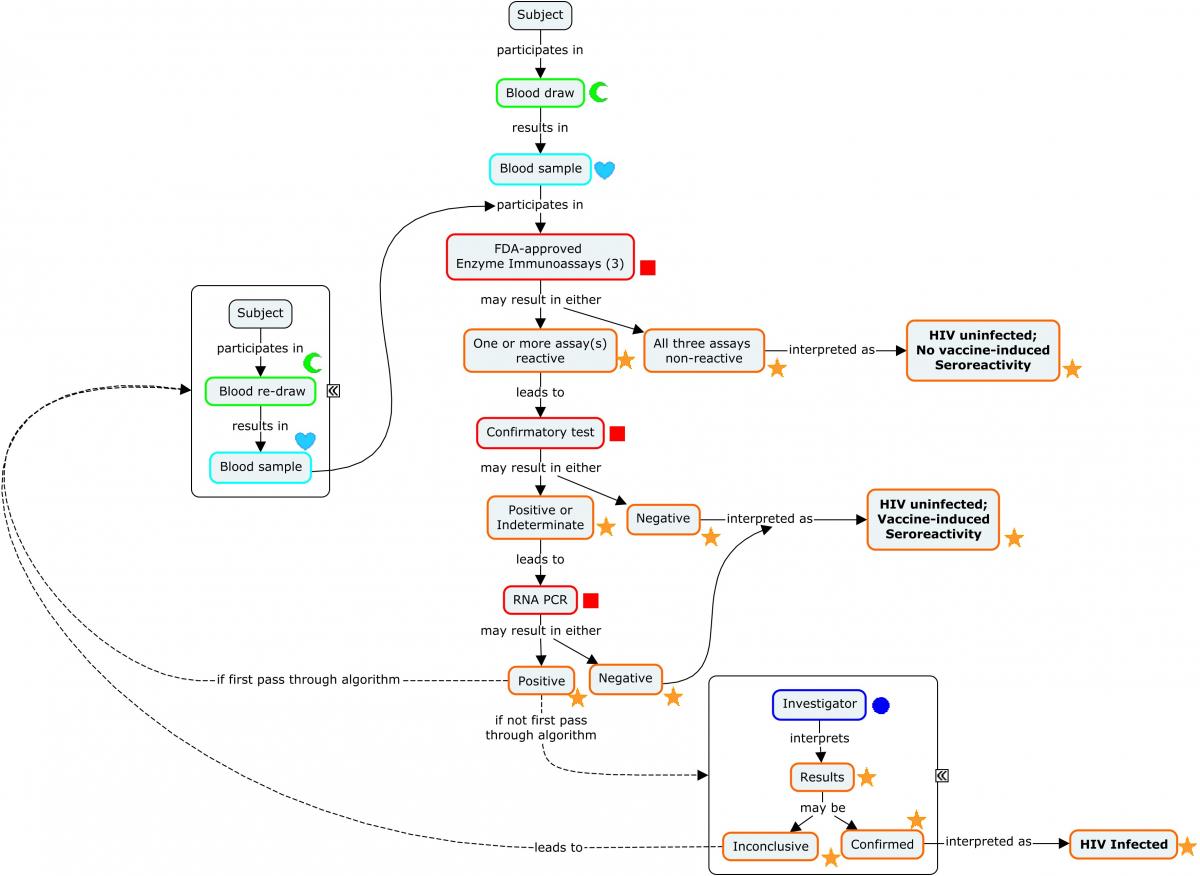
Example
In this hypothetical example, a subject participating in a vaccine trial received an HIV vaccine at the start of the study (data not shown) and was instructed to return for routine HIV testing. At the Week 6 visit, the subject tested positive for an HIV-1/2 antibody test and participated in additional confirmatory tests in accordance with the algorithm depicted in the preceding concept map. This ultimately led to the conclusion that the subject was HIV negative and had experienced vaccine-induced seroreactivity. Note that the second stage confirmatory test could also have been a western blot. In this case, the data would be handled the same: Individual antibodies tested for would be represented in ISTEST, but the method would be "WESTERN BLOT".
| Rows 1-9: | The combination of ISTESTCD/ISTEST and ISTSTDTL indicate that these tests are looking for the presence of the microbial-induced antibodies against the antigens identified in the non-standard variable ISBDAGNT. |
|---|---|
| Rows 1-3: | Show the subject was tested with 3 separate enzyme immunoassay tests for HIV and tested positive on 2 and indeterminate on the other. These tests detect the presence of antibodies against HIV-1 and -2 but do not differentiate which virus antibodies were detected. |
| Rows 4-9: | Show the results from a confirmatory immunochromatographic assay that detects antibodies to specific antigens from both HIV-2 (Rows 4-5) and HIV-1 (Rows 6-9). SPDEVID indicates that details about this assay are available in the Device Identifiers (DI) domain. Note that 2 distinct HIV-1 antibodies were detected (Rows 6 and 9). |
| Row 10: | Shows the subject is HIV-2 negative according to the assay identified in SPDEVID. The sponsor has chosen to group this interpretation record via ISGRPID to the 2 records from the assay that are relevant to HIV-2 antibody detection. |
| Row 11: | Shows the subject is HIV-1 positive according to the assay identified in SPDEVID. The sponsor has chosen to group this interpretation record via ISGRPID to the 4 records from the assay that are relevant to HIV-1 antibody detection. |
is.xpt
| Row | STUDYID | DOMAIN | USUBJID | ISSEQ | SPDEVID | ISGRPID | ISTESTCD | ISTEST | ISTSTDTL | ISORRES | ISSTRESC | ISSPEC | ISMETHOD | VISITNUM | VISIT | ISDTC | ISBDAGNT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC | IS | ABC-004 | 1 | EIA01 | MBAB | Binding Microbial-induced Antibody | DETECTION | DETECTED | DETECTED | BLOOD | EIA | 3 | WEEK 6 | 2017-05-22 | HIV-1/2 Antigen | ||
| 2 | ABC | IS | ABC-004 | 2 | EIA02 | MBAB | Binding Microbial-induced Antibody | DETECTION | INDETERMINATE | INDETERMINATE | BLOOD | EIA | 3 | WEEK 6 | 2017-05-22 | HIV-1/2 Antigen | ||
| 3 | ABC | IS | ABC-004 | 3 | EIA03 | MBAB | Binding Microbial-induced Antibody | DETECTION | DETECTED | DETECTED | BLOOD | EIA | 3 | WEEK 6 | 2017-05-22 | HIV-1/2 Antigen | ||
| 4 | ABC | IS | ABC-004 | 4 | 00105 | 1 | MBAB | Binding Microbial-induced Antibody | DETECTION | NOT DETECTED | NOT DETECTED | SERUM | IMMUNOCHROMATOGRAPHY | 3 | WEEK 6 | 2017-05-22 | HIV-2 GP36 Antigen | |
| 5 | ABC | IS | ABC-004 | 5 | 00105 | 1 | MBAB | Binding Microbial-induced Antibody | DETECTION | NOT DETECTED | NOT DETECTED | SERUM | IMMUNOCHROMATOGRAPHY | 3 | WEEK 6 | 2017-05-22 | HIV-2 GP140 Antigen | |
| 6 | ABC | IS | ABC-004 | 6 | 00105 | 2 | MBAB | Binding Microbial-induced Antibody | DETECTION | DETECTED | DETECTED | SERUM | IMMUNOCHROMATOGRAPHY | 3 | WEEK 6 | 2017-05-22 | HIV-1 P31 Antigen | |
| 7 | ABC | IS | ABC-004 | 7 | 00105 | 2 | MBAB | Binding Microbial-induced Antibody | DETECTION | NOT DETECTED | NOT DETECTED | SERUM | IMMUNOCHROMATOGRAPHY | 3 | WEEK 6 | 2017-05-22 | HIV-1 GP160 Antigen | |
| 8 | ABC | IS | ABC-004 | 8 | 00105 | 2 | MBAB | Binding Microbial-induced Antibody | DETECTION | NOT DETECTED | NOT DETECTED | SERUM | IMMUNOCHROMATOGRAPHY | 3 | WEEK 6 | 2017-05-22 | HIV-1 P24 Antigen | |
| 9 | ABC | IS | ABC-004 | 9 | 00105 | 2 | MBAB | Binding Microbial-induced Antibody | DETECTION | DETECTED | DETECTED | SERUM | IMMUNOCHROMATOGRAPHY | 3 | WEEK 6 | 2017-05-22 | HIV-1 GP41 Antigen | |
| 10 | ABC | IS | ABC-004 | 10 | 00105 | 1 | HIV2SR | HIV-2 Seroreactivity | INTERPRETATION | NEGATIVE | NEGATIVE | SERUM | IMMUNOCHROMATOGRAPHY | 3 | WEEK 6 | 2017-05-22 | ||
| 11 | ABC | IS | ABC-004 | 11 | 00105 | 2 | HIV1SR | HIV-1 Seroreactivity | INTERPRETATION | POSITIVE | POSITIVE | SERUM | IMMUNOCHROMATOGRAPHY | 3 | WEEK 6 | 2017-05-22 |
IS NSV Metadata
| Variable | Label | Type | Role | Origin |
|---|---|---|---|---|
| ISBDAGNT | Binding Agent | text | Non-Standard Record Qualifier | CRF |
The final confirmatory test uses PCR to detect HIV RNA in the subject sample. This example shows that HIV RNA was not detected in the sample, and the subject is therefore HIV negative. Because this assay is not based on immune response, it is represented in the Microbiology Specimen (MB) domain rather than the Immunogenicity Specimen Assessments (IS) domain.
mb.xpt
| Row | STUDYID | DOMAIN | USUBJID | MBSEQ | MBTESTCD | MBTEST | MBTSTDTL | MBORRES | MBSTRESC | MBSPEC | MBMETHOD | VISITNUM | VISIT | MBDTC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC | MB | ABC-004 | 1 | HIV1RNA | HIV-1 RNA | DETECTION | NOT DETECTED | NOT DETECTED | BLOOD | QUANTITATIVE REVERSE TRANSCRIPTASE POLYMERASE CHAIN REACTION | 3 | WEEK 6 | 2017-05-22 |
Based on the fact that no viral RNA was found in the subject sample and the subject was not taking prophylactic antiretroviral therapy (prohibited by protocol), the investigator concluded the subject had developed vaccine-induced seroreactivity. This event was recorded and represented in the Clinical Events (CE) domain.
ce.xpt
| Row | STUDYID | DOMAIN | USUBJID | CESEQ | CETERM | VISITNUM | VISIT | CEDTC |
|---|---|---|---|---|---|---|---|---|
| 1 | ABC | CE | ABC-004 | 1 | Vaccine-induced seroreactivity | 3 | WEEK 6 | 2017-05-22 |
Identifying information about the assay devices is represented in the Device Identifiers (DI) domain.
| Rows 1-3: | Show the device type, manufacturer, and trade name of the HIV EIA diagnostic test identified as SPDEVID="EIA01". |
|---|---|
| Rows 4-6: | Show the device type, manufacturer, and trade name of the HIV EIA diagnostic test identified as SPDEVID="EIA02". |
| Rows 7-9: | Show the device type, manufacturer, and trade name of the HIV EIA diagnostic test identified as SPDEVID="EIA03". |
| Rows 10-12: | Show the device type, manufacturer, and trade name of the confirmatory supplemental assay identified as SPDEVID="00105". |
di.xpt
| Row | STUDYID | DOMAIN | SPDEVID | DISEQ | DIPARMCD | DIPARM | DIVAL |
|---|---|---|---|---|---|---|---|
| 1 | ABC | DI | EIA01 | 1 | DEVTYPE | Device Type | HIV1/2 EIA Kit |
| 2 | ABC | DI | EIA01 | 2 | MANUF | Manufacturer | Acme |
| 3 | ABC | DI | EIA01 | 3 | TRADENAM | Trade Name | Immunowise XR |
| 4 | ABC | DI | EIA02 | 1 | DEVTYPE | Device Type | HIV1/2 EIA Kit |
| 5 | ABC | DI | EIA02 | 2 | MANUF | Manufacturer | EnzyPro |
| 6 | ABC | DI | EIA02 | 3 | TRADENAM | Trade Name | DxMate HIV |
| 7 | ABC | DI | EIA03 | 1 | DEVTYPE | Device Type | HIV1/2 EIA Kit |
| 8 | ABC | DI | EIA03 | 2 | MANUF | Manufacturer | XCelerate Dx |
| 9 | ABC | DI | EIA03 | 3 | TRADENAM | Trade Name | XCel-RNA |
| 10 | ABC | DI | 00105 | 1 | DEVTYPE | Device Type | HIV supplemental assay system |
| 11 | ABC | DI | 00105 | 2 | MANUF | Manufacturer | Bio-Rad |
| 12 | ABC | DI | 00105 | 3 | TRADENAM | Trade Name | Geenius HIV 1/2 Supplemental Assay System |
There is no RELREC dataset included as part of this example. The sponsor could have used this dataset to create a link from the records in MB and IS with the diagnosis record in CE. For more information on creating RELREC datasets, see SDTMIG Section 8.2, Relating Peer Records .
7 Disease Assessments
This section generally provides information on data used to evaluate how a subject is progressing over the course of a study. These data are typically collected multiple times during the study. However, this section includes an example (Section 7.1, CD4 Counts) illustrating the representation of both pre- and on-study CD4 counts. While pre-study CD4 counts would generally be represented in Section 3, Subject and Disease Characteristics, for clarity they are shown together in this section with on-study assessments.
7.1 CD4 Counts
CD4 counts are an important lab result to collect for HIV-infected people. In addition to determining CD4 counts during a study, subjects are often asked about their nadir (i.e., lowest) CD4 count. The source of the nadir CD4 count may be a subject's medical records, but may also be based on subject recall. Since a subject may not be able to recall an exact number, nadir CD4 count may be collected using ranges, rather than an exact value.
Example
The representation of both historical and on-study CD4 counts is shown for one subject at a screening visit. A sample was collected, the CD4 count was determined and electronically reported by a central lab (i.e., a case report form was not completed). The subject was also asked several questions about lifetime nadir (lowest) CD4 count and the responses were recorded on the case report form shown below.
The Nadir CD4 CRF was used to solicit information about a subject's historical lowest CD4 count obtained by medical records and subject recall. In this example CRF, the following syntax was used to create denormalized CDASH variable names: SPONSOR DEFINED NAME_TESTCD_ROOT VARIABLE.
Historical Nadir CD4 CRF
| Indicate if there are any CD4+ absolute counts with source documentation available. If Yes, include the appropriate details where indicated on the CRF. | Were there CD4+ absolute counts with source documentation?
DOC_CD4_LBYN Not submitted |
<From NY codelist> |
|---|---|---|
| Record the lowest CD4+ absolute count in cells/uL for which there is source documentation. |
DOC_CD4_LBORRES LBORRES | _____ |
|
DOC_CD4_LBORRESU LBORRESU Pre-populated | 10^6/L
<From UNIT codelist> | |
| Record the date of specimen collection according to the source documentation. |
DOC_CD4_LBDAT LBDTC | _ _ / _ _ _ / _ _ _ _ |
| Indicate whether the subject recalls a different lowest CD4+ absolute count for which there is no source documentation. If Yes, include the appropriate details where indicated on the CRF. | Does the subject recall a lower CD4+ absolute count than is in the source documentation?
RECALL_CD4_LBYN Not submitted |
<From NY codelist> |
| Record the subject-recalled lowest CD4+ count in cells/uL. | Result
RECALL_CD4_LBORRES LBORRES |
|
|
RECALL_CD4_LBORRESU LBORRESU Pre-populated | 10^6/L
<From UNIT codelist> |
CRF Metadata
It is important to be able to distinguish pre-study nadir CD4 counts from CD4 counts collected during the study. In this example, nadir CD4 values can be recognized by:
- Timing variables that indicate that the finding was pre-study
- If the date of the nadir value is available, LBDY will be present and be negative
- If a date of the nadir is not available, the relative timing variable LBSTRF have the value of "BEFORE", showing that the test was conducted before the study reference period
- The presence of the variable "Lab Category" (LBCAT), with a value of "HEMATOLOGY HISTORY"
- The presence of the NSV "Source of Data" (LBSOURCE), with values such as "MEDICAL RECORD" or "SUBJECT RECALL"
In this example, the visit variables are populated for all the data collected at the screening visit. The inclusion of the visit variables in the records for nadir CD4 values is intended to represent the fact that the data were collected at this visit, not that the tests were conducted at the screening visit.
Programs used to derive Study Visit (SV) datasets from visit dates in records for collected data should be written so as not to include pre-study dates for assessments collected at screening. One strategy may be to exclude dates prior to the date of informed consent.
This example shows data for a study subject. If the data were about an associated person, the data would be represented in the APLB domain and the dataset would include APID, RSUBJID, and SREL, rather than USUBJID.
| Row 1: | Shows the CD4 count of the subject on Study Day 1, as reported by Laboratory A. Note that the name of the laboratory vendor used was represented in LBNAM, |
|---|---|
| Row 2: | Shows the nadir CD4 count before the start of the study, as determined by a review of the available medical records. LBDTC represents the specimen collection date associated with the nadir CD4 , which in this case was 989 days before the start of the study.. The non-standard qualifier LBCOLSRT indicates that this value represents a nadir CD4 count ("LOWEST"). The non-standard qualifier LBSOURCE provides the information that this value came from the subject's medical record. The variable LBEVINTX is populated with "LIFETIME" to indicate the evaluation period. |
| Row 3: | Shows that the subject recalled a different nadir CD4 count than that provided by the source documentation. The subject's recalled nadir CD4 count was reported as a range, rather than a value, and the source was "SUBJECT RECALL" (LBSOURCE). The variable LBDTC is not populated, as the date of specimen collection for the subject's recalled CD4 test was not collected. Instead, LBSTRF (Start Relative to Reference Period) is populated with the value of "BEFORE" to show that this is a pre-study value. The non-standard qualifier LBCOLSRT indicates that this value represents a nadir CD4 count ("LOWEST" ). The variable LBEVINTX is populated with "LIFETIME" to indicate the evaluation period. |
lb.xpt
| Row | STUDYID | DOMAIN | USUBJID | LBSEQ | LBREFID | LBTESTCD | LBTEST | LBCAT | LBORRES | LBORRESU | LBSTRESC | LBSTRESN | LBSTRESU | LBNAM | VISITNUM | VISIT | VISITDY | LBDY | LBDTC | LBSTRF | LBEVINTX | LBCOLSRT | LBSOURCE | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HIV-01 | LB | HIV-01-001 | 1 | 12342 | CD4 | CD4 | HEMATOLOGY | 220 | 10^6/L | 220 | 220 | 10^6/L | LAB A | 1 | SCREENING | 1 | 1 | 2015-03-04 | |||||
| 2 | HIV-01 | LB | HIV-01-001 | 2 | 79343 | CD4 | CD4 | HEMATOLOGY HISTORY | 210 | 10^6/L | 210 | 210 | 10^6/L | 1 | SCREENING | 1 | -989 | 2012-06-18 | LIFETIME | LOWEST | MEDICAL RECORD- | |||
| 3 | HIV-01 | LB | HIV-01-001 | 3 | CD4 | CD4 | HEMATOLOGY HISTORY | 51-199 | 10^6/L | 51-199 | 10^6/L | 1 | SCREENING | 1 | BEFORE | LIFETIME | LOWEST | SUBJECT RECALL |
LB NSV Metadata
| Variable | Label | Type | Role | Origin |
|---|---|---|---|---|
| LBCOLSRT | Collected Summary Result Type | text | Non-Standard Record Qualifier | CRF |
| LBSOURCE | Source of Data | text | Non-Standard Record Qualifier | CRF |
7.2 HIV RNA Detection and Viral Load
Antiretroviral therapy can successfully reduce morbidity and mortality associated with HIV infection and can also reduce transmission of the virus. The primary measure of effective ART in HIV-positive subjects is viral suppression as indicated by a reduction in viral load over time.
Example
This example follows 1 subject through a series of viral load assessments. By the visit at Week 48, the viral load had dropped below the limit of detection. Note the use of MBTSTDTL to indicate that test of the analyte "HIV-1 RNA" was a measurement of viral load. In this example, the sponsor populated MBLOINC because it was provided by the central lab.
| Row 1: | Shows the HIV-1 viral load was measured as 496700 copies/mL at the baseline visit. |
|---|---|
| Rows 2-3: | Show that the HIV-1 viral load continued to drop between Weeks 4 and 24. |
| Row 4: | Shows that HIV-1 RNA was detected in the subject's sample at the Week 48 visit, but the viral load was below the lower limit of quantitation. Because this was an attempt to measure viral load, MBTSTDTL="VIRAL LOAD", even though the assessment did not result in a quantifiable result. |
| Row 5: | Shows that the subject's HIV-1 viral load had dropped below the limit of detection by Week 72. |
mb.xpt
| Row | STUDYID | DOMAIN | USUBJID | SPDEVID | MBSEQ | MBREFID | MBTESTCD | MBTEST | MBTSTDTL | MBORRES | MBORRESU | MBSTRESC | MBSTRESN | MBSTRESU | MBLOINC | MBSPEC | MBMETHOD | MBLLOQ | MBBLFL | VISITNUM | VISIT | MBDTC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC | MB | ABC-001 | RT-qPCR01 | 1 | 001-02 | HIV1RNA | HIV-1 RNA | VIRAL LOAD | 496700 | copies/mL | 496700 | 496700 | copies/mL | 70241-5 | PLASMA | QUANTITATIVE REVERSE TRANSCRIPTASE POLYMERASE CHAIN REACTION | 20 | Y | 2 | BASELINE | 2016-05-19 |
| 2 | ABC | MB | ABC-001 | RT-qPCR01 | 2 | 001-03 | HIV1RNA | HIV-1 RNA | VIRAL LOAD | 19400 | copies/mL | 19400 | 19400 | copies/mL | 70241-5 | PLASMA | QUANTITATIVE REVERSE TRANSCRIPTASE POLYMERASE CHAIN REACTION | 20 | 3 | WEEK 4 | 2016-06-10 | |
| 3 | ABC | MB | ABC-001 | RT-qPCR02 | 3 | 001-04 | HIV1RNA | HIV-1 RNA | VIRAL LOAD | 79 | copies/mL | 79 | 79 | copies/mL | 70241-5 | PLASMA | QUANTITATIVE REVERSE TRANSCRIPTASE POLYMERASE CHAIN REACTION | 20 | 4 | WEEK 24 | 2016-11-08 | |
| 4 | ABC | MB | ABC-001 | RT-qPCR02 | 4 | 001-05 | HIV1RNA | HIV-1 RNA | VIRAL LOAD | TARGET DETECTED, BELOW LLOQ | TARGET DETECTED, BELOW LLOQ | PLASMA | QUANTITATIVE REVERSE TRANSCRIPTASE POLYMERASE CHAIN REACTION | 20 | 5 | WEEK 48 | 2017-04-06 | |||||
| 5 | ABC | MB | ABC-001 | RT-qPCR03 | 5 | 001-06 | HIV1RNA | HIV-1 RNA | VIRAL LOAD | TARGET NOT DETECTED | TARGET NOT DETECTED | PLASMA | QUANTITATIVE REVERSE TRANSCRIPTASE POLYMERASE CHAIN REACTION | 20 | 6 | WEEK 72 | 2017-10-13 |
Properties of the assays used to determine viral load are represented in the device domains. In the example below, the lot numbers of the assays used changed, and the sponsor chose to keep track of this parameter. Had the lot number not been changed or had not been of interest, a single SPDEVID could have been used to link to a single set of parameters in the Device Identifiers (DI) dataset.
| Rows 1-4: | Show the device type, manufacturer, trade name, and lot number for the RT-qPCR kit identifed by SPDEVID="RT-qPCR01". |
|---|---|
| Rows 5-8: | Show the device type, manufacturer, trade name, and lot number for the RT-qPCR kit identifed by SPDEVID="RT-qPCR02". |
| Rows 9-12: | Show the device type, manufacturer, trade name, and lot number for the RT-qPCR kit identifed by SPDEVID="RT-qPCR03". |
di.xpt
| Row | STUDYID | DOMAIN | SPDEVID | DISEQ | DIPARMCD | DIPARM | DIVAL |
|---|---|---|---|---|---|---|---|
| 1 | ABC | DI | RT-qPCR01 | 1 | DEVTYPE | Device Type | RT-qPCR kit |
| 2 | ABC | DI | RT-qPCR01 | 2 | MANUF | Manufacturer | Acme |
| 3 | ABC | DI | RT-qPCR01 | 3 | TRADENAM | Trade Name | DetectPRO |
| 4 | ABC | DI | RT-qPCR01 | 4 | LOT | Lot Number | 20160202013 |
| 5 | ABC | DI | RT-qPCR02 | 1 | DEVTYPE | Device Type | RT-qPCR kit |
| 6 | ABC | DI | RT-qPCR02 | 2 | MANUF | Manufacturer | Acme |
| 7 | ABC | DI | RT-qPCR02 | 3 | TRADENAM | Trade Name | DetectPRO |
| 8 | ABC | DI | RT-qPCR02 | 4 | LOTNUM | Lot Number | 20161101004 |
| 9 | ABC | DI | RT-qPCR03 | 1 | DEVTYPE | Device Type | RT-qPCR kit |
| 10 | ABC | DI | RT-qPCR03 | 2 | MANUF | Manufacturer | Acme |
| 11 | ABC | DI | RT-qPCR03 | 3 | TRADENAM | Trade Name | DetectPRO |
| 12 | ABC | DI | RT-qPCR03 | 4 | LOTNUM | Lot Number | 201708101009 |
7.3 Drug Susceptibility
This section covers phenotypic and genotypic resistance testing as it pertains to HIV. The following concept map illustrates these concepts at a high level that is not specific to HIV.
Concept Map. Drug Sensitivity Testing
7.3.1 Phenotypic Drug Sensitivity Testing
Phenotypic drug sensitivity testing involves culturing the virus in a cell line that is permissive to infection. This topic is covered by the Virology Therapeutic Area User Guide v2.1 (https://www.cdisc.org/standards/therapeutic-areas/virology) and no additional examples are shown here. Users should refer to the Resistance Testing section of that guide for more information and illustrative examples on modeling data from these assays.
7.3.2 Genetic Resistance Calculators and Interpretations
Genetic resistance calculators provide estimates of drug resistance or susceptibility based on what is known about the genetic variations present in specific genes within the HIV isolated from a subject sample.
Example
First, the mutations data are represented in the Pharmacogenomics/Genetics Findings (PF) domain. The example below shows what a subset of mutations might look like for mutations reported as amino acid changes in the HIV-1 reverse transcriptase gene. In a study setting, these results could be reported as nucleotide, amino acid, or codon changes and there could be many records documenting mutations across multiple genes. Every change from the consensus (reference) sequence should be represented, whether or not it contributes to the overall resistance to a drug. Note that NHOID is used to clarify that these genetic variations are viral genetic results, rather than genetic results for the subject. PFSTRESC shows the HGVS nomenclature for each variant. Row 10 shows a case where there was a mixed population of viruses with respect to the same location: those with either a change to F (Phe) or a change to V (Val), as indicated in PFORRES and PFSTRESC.
pf.xpt
| Row | STUDYID | DOMAIN | USUBJID | NHOID | PFSEQ | PFTESTCD | PFTEST | PFGENRI | PFGENTYP | PFREFSEQ | PFORRES | PFORREF | PFGENLOC | PFSTRESC | PFMETHOD | VISITNUM | PFDTC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HIV01 | PF | HIV01-101 | HIV-1 | 1 | GENVAR | Genetic Variation | REVERSE TRANSCRIPTASE | PROTEIN | CONSENSUS B | I | V | 60 | p.(Val60Ile) | NEXT GENERATION SEQUENCING | 2 | 2018-07-31 |
| 2 | HIV01 | PF | HIV01-101 | HIV-1 | 2 | GENVAR | Genetic Variation | REVERSE TRANSCRIPTASE | PROTEIN | CONSENSUS B | R | K | 65 | p.(Lys65Arg) | NEXT GENERATION SEQUENCING | 2 | 2018-07-31 |
| 3 | HIV01 | PF | HIV01-101 | HIV-1 | 3 | GENVAR | Genetic Variation | REVERSE TRANSCRIPTASE | PROTEIN | CONSENSUS B | N | D | 67 | p.(Asp67Asn) | NEXT GENERATION SEQUENCING | 2 | 2018-07-31 |
| 4 | HIV01 | PF | HIV01-101 | HIV-1 | 4 | GENVAR | Genetic Variation | REVERSE TRANSCRIPTASE | PROTEIN | CONSENSUS B | N | T | 69 | p.(Thr69Asn) | NEXT GENERATION SEQUENCING | 2 | 2018-07-31 |
| 5 | HIV01 | PF | HIV01-101 | HIV-1 | 5 | GENVAR | Genetic Variation | REVERSE TRANSCRIPTASE | PROTEIN | CONSENSUS B | R | K | 70 | p.(Lys70Arg) | NEXT GENERATION SEQUENCING | 2 | 2018-07-31 |
| 6 | HIV01 | PF | HIV01-101 | HIV-1 | 6 | GENVAR | Genetic Variation | REVERSE TRANSCRIPTASE | PROTEIN | CONSENSUS B | E | K | 122 | p.(Lys122Glu) | NEXT GENERATION SEQUENCING | 2 | 2018-07-31 |
| 7 | HIV01 | PF | HIV01-101 | HIV-1 | 7 | GENVAR | Genetic Variation | REVERSE TRANSCRIPTASE | PROTEIN | CONSENSUS B | K | T | 139 | p.(Thr139Lys) | NEXT GENERATION SEQUENCING | 2 | 2018-07-31 |
| 8 | HIV01 | PF | HIV01-101 | HIV-1 | 8 | GENVAR | Genetic Variation | REVERSE TRANSCRIPTASE | PROTEIN | CONSENSUS B | V | M | 184 | p.(Met184Val) | NEXT GENERATION SEQUENCING | 2 | 2018-07-31 |
| 9 | HIV01 | PF | HIV01-101 | HIV-1 | 9 | GENVAR | Genetic Variation | REVERSE TRANSCRIPTASE | PROTEIN | CONSENSUS B | A | T | 200 | p.(Thr200Ala) | NEXT GENERATION SEQUENCING | 2 | 2018-07-31 |
| 10 | HIV01 | PF | HIV01-101 | HIV-1 | 10 | GENVAR | Genetic Variation | REVERSE TRANSCRIPTASE | PROTEIN | CONSENSUS B | FV | T | 215 | p.(Thr215Phe^Val) | NEXT GENERATION SEQUENCING | 2 | 2018-07-31 |
| 11 | HIV01 | PF | HIV01-101 | HIV-1 | 11 | GENVAR | Genetic Variation | REVERSE TRANSCRIPTASE | PROTEIN | CONSENSUS B | C | K | 219 | p.(Lys219Cys) | NEXT GENERATION SEQUENCING | 2 | 2018-07-31 |
| 12 | HIV01 | PF | HIV01-101 | HIV-1 | 12 | GENVAR | Genetic Variation | REVERSE TRANSCRIPTASE | PROTEIN | CONSENSUS B | H | Q | 334 | p.(Gln334His) | NEXT GENERATION SEQUENCING | 2 | 2018-07-31 |
| 13 | HIV01 | PF | HIV01-101 | HIV-1 | 13 | GENVAR | Genetic Variation | REVERSE TRANSCRIPTASE | PROTEIN | CONSENSUS B | D | G | 335 | p.(Gly335Asp) | NEXT GENERATION SEQUENCING | 2 | 2018-07-31 |
| 14 | HIV01 | PF | HIV01-101 | HIV-1 | 14 | GENVAR | Genetic Variation | REVERSE TRANSCRIPTASE | PROTEIN | CONSENSUS B | I | N | 348 | p.(Asn348Ile) | NEXT GENERATION SEQUENCING | 2 | 2018-07-31 |
Next, the summary report generated by the resistance calculator is represented as records in the Microbiology Susceptibility (MS) domain. Only 5 of the 14 changes in the reverse transcriptase gene represented in PF above were scored by the resistance calculator algorithm for tenofovir, the drug named in MSAGENT. A total of 6 mutations were scored for the nucleaoside reverse transicriptase inhibitor drug class. The 8 remaining unscored variants shown above presumably do not affect resistance to the drug. In this example, the sponsor chose to represent the list of scored mutations relevant to each record in the non-standard variable MSSCRMUT to match how the data were received in the output of the algorithm. RELREC could have also been used to link the resistance summary conclusions in MS with the mutations in PF.
| Row 1: |
Shows the resistance calculator raw score for tenofovir, based on the mutations present. MSANMETH identifies the Stanford resistance calculator as the source of this score. The non-standard variable MSSCRMUT lists all the mutations that the algorithm scored for tenofovir. MSGRPID is used to group this score record with its interpretation in Row 2. |
|---|---|
| Row 2: | Shows the resistance level (Intermediate Resistance) to tenofovir as determined by the Stanford resistance calculator based on the genetic variants listed in the non-standard variable MSSCRMUT. MSGRPID groups this record with the corresponding score shown in Row 1. |
| Row 3: | Shows that the Stanford resistance calculator algorithm identified the 6 mutations shown in the NSV MSSCRMUT as conferring some level of resistance to reverse transcriptase inhibitors at the class level. Note that MSAGENT="NUCLEOSIDE REVERSE TRANSCRIPTASE INHIBITORS" to indicate that this finding pertains to a drug class as opposed to an individual drug. There is no associated score or resistance level associated with this finding, as it may vary by individual drug. This record instead serves to indicate that using this class of drug with this subject may be less effective. |
ms.xpt
| Row | STUDYID | DOMAIN | USUBJID | NHOID | MSSEQ | MSGRPID | MSREFID | MSTESTCD | MSTEST | MSTSTDTL | MSAGENT | MSCAT | MSORRES | MSSTRESC | MSSTRESN | MSANMETH | VISITNUM | MSSCRMUT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HIV01 | MS | HIV01-101 | HIV-1 | 1 | 1 | 101-001 | MICROSUS | Microbial Susceptibility | SCORE | TENOFOVIR | GENETIC RESISTANCE | 35 | 35 | 35 | STANFORD RESISTANCE CALCULATOR V8.4 | 2 | D67N,K70R,M184V,T215FV,K219C | |
| 2 | HIV01 | MS | HIV01-101 | HIV-1 | 2 | 1 | 101-001 | MICROSUS | Microbial Susceptibility | INTERPRETATION | TENOFOVIR | GENETIC RESISTANCE | Intermediate resistance | INTERMEDIATE RESISTANCE | STANFORD RESISTANCE CALCULATOR V8.4 | 2 | D67N,K70R,M184V,T215FV,K219C | ||
| 3 | HIV01 | MS | HIV01-101 | HIV-1 | 3 | 101-001 | SUSMUIND | Susceptibility Score Mutations Indicator | NUCLEOSIDE REVERSE TRANSCRIPTASE INHIBITORS | GENETIC RESISTANCE | Yes | Y | STANFORD RESISTANCE CALCULATOR V8.4 | 2 | D67N,K70R,M184V,T215FV,K219C, N348I |
MS NSV Metadata
| Variable | Label | Type | Role | Origin |
|---|---|---|---|---|
| MSSCRMUT |
List of All Mutations Scored | text | Non-Standard Record Qualifier | eDT |
While not strictly a drug resistance calculator, an HIV-1 sequence may undergo expert interpretation. For example, the International Antiviral Association–USA (IAS–USA) Drug Resistance Mutations Group is an independent, volunteer panel of experts charged with delivering accurate, unbiased, and evidence-based information on drug resistance-associated mutations for HIV clinical practitioners.[12] This group reviews new data on HIV drug resistance to maintain a current list of mutations associated with clinical resistance to HIV-1 . This list includes mutations that may contribute to a reduced virologic response to drugs that have been approved by the FDA, as well as any drugs available in expanded access programs. While expert resistance interpretations such as IAS–USA may not provide a measure of the level of drug susceptibility, they do provide an indicator that a shift in drug susceptibility has occurred. Example 2 is based on the IAS–USA interpretation of the same mutations shown in the pf.xpt example in Example 1.
Example
This example shows resistance information (based on mutations present in a subject sample) as it would be identified by the IAS–USA expert panel. As in the case of data from the Stanford algorithm, these data are represented in MS. In this example, the sponsor chose to represent the flagged mutations present to match how the data were received. RELREC could have also been used to link the resistance summary conclusions in MS with the mutations in PF.
| Row 1: |
Indicates that one of the mutations present in the subject's HIV sample was flagged by the IAS-USA expert panel as being associated with susceptibility to tenofovir. The non-standard variable MSFLMUT shows the specific mutation. The NSV MSREFUN shows that the IAS-USA 2017 expert panel publication refers investigators to a "user note" pertaining to this mutation. |
|---|---|
| Row 2: |
Shows that another set of mutations were flagged by the IAS-USA expert panel for conferring resistance to reverse transcriptase inhibitors at the drug class level. Note that MSAGENT="NUCLEOSIDE REVERSE TRANSCRIPTASE INHIBITORS" to indicate that this finding pertains to a drug class as opposed to an individual drug. Additionally, a user note from the IAS-USA 2017 list was referenced for further information. |
ms.xpt
| Row | STUDYID | DOMAIN | USUBJID | NHOID | MSSEQ | MSGRPID | MSREFID | MSTESTCD | MSTEST | MSAGENT | MSCAT | MSORRES | MSSTRESC | MSANMETH | VISITNUM | MSFLMUT | MSREFUN | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HIV01 | MS | HIV01-101 | HIV-1 | 1 | 1 | 101-001 |
FLMUTIND | Flagged Mutations Present Indicator | TENOFOVIR | GENETIC RESISTANCE | Yes | Y | IAS-USA 201708 | 2 | K65R | USERNOTE_I | |
| 2 | HIV01 | MS | HIV01-101 | HIV-1 | 3 | 2 | 101-001 | FLMUTIND | Flagged Mutations Present Indicator | NUCLEOSIDE REVERSE TRANSCRIPTASE INHIBITORS | GENETIC RESISTANCE | Yes | Y | IAS-USA 201708 | 2 | K65R, D67N, K70R, T215FV | USERNOTE_L |
MS NSV Metadata
| Variable | Label | Type | Role | Origin |
|---|---|---|---|---|
| MSFLMUT | List of All Mutations Flagged | text | Non-Standard Record Qualifier | eDT |
| MSREFUN | Referenced User Note | text | Non-Standard Record Qualifier | eDT |
7.4 DXA Scans
Dual-energy X-Ray Absorptiometry (DEXA or DXA) is an imaging technique used to measure bone mineral density (BMD). The purpose of conducting DXA scan assessments in HIV is twofold:
- antiretroviral therapy (ART) is associated with a 2-6% decline in BMD by year 2 post-initiation of therapy, and
- the virus itself may cause loss of BMD.
Prevalence of osteoporosis in HIV-infected individuals may be as high as 3 times that of HIV-negative matched controls, with 30-70% greater rate of fractures.[13] In addition to measuring absolute BMD, the image analysis provided by the scanner system software also computes Z scores and T scores to show how a subject compares to the general population. The Z score is the number of standard deviations the observed result falls either above (positive Z score) or below (negative Z score) the mean observed value for population-matched controls. The T score, in BMD assessment at least, is the Z score when the control population is limited to healthy 30-year-olds.
Example
This example shows data associated with a series of DXA scans performed first at a screening visit, then every 48 weeks for 2 more visits. The measurements focus on the lumbar spine and the hip (the femoral neck), as these areas support much of the body's weight.
First, the procedure may be represented in the Procedures (PR) domain. One common reason for representing procedures in the PR domain is to represent the responses to questions about whether a planned procedure was performed.
pr.xpt
| Row | STUDYID | DOMAIN | USUBJID | SPDEVID | PRSEQ | PRTRT | PRPRESP | PROCCUR | VISITNUM | VISIT | PRSTDTC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC | PR | ABC-01-101 | DX13 | 1 | DXA SCAN | Y | Y | 1 | SCREENING | 2015-08-04 |
| 2 | ABC | PR | ABC-01-101 | DX13 | 2 | DXA SCAN | Y | Y | 6 | WEEK 48 | 2016-06-28 |
| 3 | ABC | PR | ABC-01-101 | DX13 | 3 | DXA SCAN | Y | Y | 12 | WEEK 96 | 2017-06-02 |
The results from the scan are represented in the Musculoskeletal Findings (MK) domain. The scan results represented include bone mineral density (BMD), Z score, and T score for each of 2 regions across all visits. Note the gradual reduction in BMD (and associated Z scores and T scores) over the course of 3 visits spanning approximately 2 years.
| Rows 1-3: | Show the subject's bone mineral density, Z score, and T score from a DXA scan of the lumbar spine at the screening visit. |
|---|---|
| Rows 4-6: | Show the subject's bone mineral density, Z score, and T score from a DXA scan of the left femoral neck at the screening visit. |
| Rows 7-12: | Show the subject's bone mineral density, Z score, and T score from a DXA scan of the same 2 regions at Week 48. |
| Rows 13-18: | Show the subject's bone mineral density, Z score, and T score from a DXA scan of the same 2 regions at Week 96. |
mk.xpt
| Row | STUDYID | DOMAIN | USUBJID | SPDEVID | MKSEQ | MKTESTCD | MKTEST | MKORRES | MKORRESU | MKSTRESC | MKSTRESN | MKSTRESU | MKLOC | MKLAT | MKMETHOD | VISITNUM | VISIT | MKDTC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC | MK | ABC-01-101 | DX13 | 1 | BMD | Bone Mineral Density | 1.126 | g/cm2 | 1.126 | 1.126 | g/cm2 | SPINE, LUMBAR | DXA SCAN | 1 | SCREENING | 2015-08-04 | |
| 2 | ABC | MK | ABC-01-101 | DX13 | 2 | BMDZ | Bone Mineral Density Z Score | 0.401 | 0.401 | 0.401 | SPINE, LUMBAR | DXA SCAN | 1 | SCREENING | 2015-08-04 | |||
| 3 | ABC | MK | ABC-01-101 | DX13 | 3 | BMDT | Bone Mineral Density T Score | 0.011 | 0.011 | 0.011 | SPINE, LUMBAR | DXA SCAN | 1 | SCREENING | 2015-08-04 | |||
| 4 | ABC | MK | ABC-01-101 | DX13 | 4 | BMD | Bone Mineral Density | 1.131 | g/cm2 | 1.131 | 1.131 | g/cm2 | FEMORAL NECK | LEFT | DXA SCAN | 1 | SCREENING | 2015-08-04 |
| 5 | ABC | MK | ABC-01-101 | DX13 | 5 | BMDZ | Bone Mineral Density Z Score | 0.201 | 0.201 | 0.201 | FEMORAL NECK | LEFT | DXA SCAN | 1 | SCREENING | 2015-08-04 | ||
| 6 | ABC | MK | ABC-01-101 | DX13 | 6 | BMDT | Bone Mineral Density T Score | 0.019 | 0.019 | 0.019 | FEMORAL NECK | LEFT | DXA SCAN | 1 | SCREENING | 2015-08-04 | ||
| 7 | ABC | MK | ABC-01-101 | DX13 | 7 | BMD | Bone Mineral Density | 1.112 | g/cm2 | 1.112 | 1.112 | g/cm2 | SPINE, LUMBAR | DXA SCAN | 6 | WEEK 48 | 2016-06-28 | |
| 8 | ABC | MK | ABC-01-101 | DX13 | 8 | BMDZ | Bone Mineral Density Z Score | 0.011 | 0.011 | 0.011 | SPINE, LUMBAR | DXA SCAN | 6 | WEEK 48 | 2016-06-28 | |||
| 9 | ABC | MK | ABC-01-101 | DX13 | 9 | BMDT | Bone Mineral Density T-Score | -0.509 | -0.509 | -0.509 | SPINE, LUMBAR | DXA SCAN | 6 | WEEK 48 | 2016-06-28 | |||
| 10 | ABC | MK | ABC-01-101 | DX13 | 10 | BMD | Bone Mineral Density | 1.108 | g/cm2 | 1.108 | 1.108 | g/cm2 | FEMORAL NECK | LEFT | DXA SCAN | 6 | WEEK 48 | 2016-06-28 |
| 11 | ABC | MK | ABC-01-101 | DX13 | 11 | BMDZ | Bone Mineral Density Z Score | -0.011 | -0.011 | -0.011 | FEMORAL NECK | LEFT | DXA SCAN | 6 | WEEK 48 | 2016-06-28 | ||
| 12 | ABC | MK | ABC-01-101 | DX13 | 12 | BMDT | Bone Mineral Density T Score | -0.102 | -0.102 | -0.102 | FEMORAL NECK | LEFT | DXA SCAN | 6 | WEEK 48 | 2016-06-28 | ||
| 13 | ABC | MK | ABC-01-101 | DX13 | 13 | BMD | Bone Mineral Density | 1.092 | g/cm2 | 1.092 | 1.092 | g/cm2 | SPINE, LUMBAR | DXA SCAN | 12 | WEEK 96 | 2017-06-02 | |
| 14 | ABC | MK | ABC-01-101 | DX13 | 14 | BMDZ | Bone Mineral Density Z Score | -0.502 | -0.502 | -0.502 | SPINE, LUMBAR | DXA SCAN | 12 | WEEK 96 | 2017-06-02 | |||
| 15 | ABC | MK | ABC-01-101 | DX13 | 15 | BMDT | Bone Mineral Density T-Score | -0.704 | -0.704 | -0.704 | SPINE, LUMBAR | DXA SCAN | 12 | WEEK 96 | 2017-06-02 | |||
| 16 | ABC | MK | ABC-01-101 | DX13 | 16 | BMD | Bone Mineral Density | 1.103 | g/cm2 | 1.103 | 1.103 | g/cm2 | FEMORAL NECK | LEFT | DXA SCAN | 12 | WEEK 96 | 2017-06-02 |
| 17 | ABC | MK | ABC-01-101 | DX13 | 17 | BMDZ | Bone Mineral Density Z Score | -0.152 | -0.152 | -0.152 | FEMORAL NECK | LEFT | DXA SCAN | 12 | WEEK 96 | 2017-06-02 | ||
| 18 | ABC | MK | ABC-01-101 | DX13 | 18 | BMDT | Bone Mineral Density T Score | -0.313 | -0.313 | -0.313 | FEMORAL NECK | LEFT | DXA SCAN | 12 | WEEK 96 | 2017-06-02 |
The device used in the scan is identified in the PR and MK domains by the identifier SPDEVID. The Device Identifiers (DI) domain provides the identifying characteristics of this device. Capturing this information may be particularly important in imaging, where changes in the equipment used across a study can significantly affect results.
di.xpt
| Row | STUDYID | DOMAIN | SPDEVID | DISEQ | DIPARMCD | DIPARM | DIVAL |
|---|---|---|---|---|---|---|---|
| 1 | ABC | DI | DX13 | 1 | DEVTYPE | Device Type | DUAL-ENERGY X-RAY ABSORPTIOMETRY TOTAL BODY SCANNER |
| 2 | ABC | DI | DX13 | 2 | MANUF | Manufacturer | HOLOGIC |
| 3 | ABC | DI | DX13 | 3 | MODEL | Model | QDR-4500A |
The results expressed as Z scores are based on reference equations used by the device. The reference equation used is represented in the Device In Use (DU) domain, as is the version of the software running on the scanner.
du.xpt
| Row | STUDYID | DOMAIN | USUBJID | SPDEVID | DUSEQ | DUTESTCD | DUTEST | DUORRES |
|---|---|---|---|---|---|---|---|---|
| 1 | ABC | DU | ABC-01-101 | DX13 | 1 | DXAREFEQ | DXA Reference Equation | NATIONAL HEALTH AND NUTRITION EXAMINATION SURVEY (NHANES) III 2010 |
| 2 | ABC | DU | ABC-01-101 | DX13 | 2 | SFTWRVER | Software Version | 16 |
7.5 Flow Cytometry
A CDISC subteam is currently working on examples for HIV-related flow cytometry results. This work is ongoing and is expected to be published in a future version of the SDTMIG.
7.6 Questionnaires, Ratings, and Scales
eQuestionnaires, Ratings, and Scales (QRSs) are typically done repeatedly over the course of a study. These are used to evaluate how a subject is progressing, by assessing the severity of disease and improvement or worsening of specific symptoms.
QRS measures are maintained as stand-alone guides on the CDISC website (https://www.cdisc.org/foundational/qrs). The following table lists assessments that are being pursued as potential supplements as part of the development work for the TAUG-HIV. Supplements may or may not be finalized at the time of publication of this user guide, and depend on copyright approval where applicable. CDISC cannot produce supplements for copyrighted measures without the express permission of the copyright holder.
Sponsors should refer to the link above if a measure of interest is not included below, as it may have been developed for another therapeutic area; new measures are implemented on an ongoing basis by the CDISC QRS Terminology and Standards Development subteams. Table. Measures Relevant to HIV Research
| Full Name and Abbreviation | Copyright Permission Status | Supplement Status |
|---|---|---|
| HIV Treatment Satisfaction Questionnaire Status Version 14+ (HIVTSQs) | To be requested | |
| HIV Treatment Satisfaction Questionnaire Change Version 14+ (HIVTSQc) | To be requested | |
| Perception of Injection (PIN) | More information needed | |
| HIV/AIDS-Targeted Quality of Life (HATQoL) | To be requested | |
| ACCEPT | More information needed | |
| EQ5D/EQ5D-5L | Granted | Done |
| CDC Classification System for HIV-Infected Adults and Adolescents | Granted | Done |
| World Health Organization HIV Classification System | Granted | Done |
| Symptoms Distress Module | More information needed | |
| Apgar Score | Requested | |
| Edinburgh Postnatal Depression Study | Granted | Supplement in progress |
| Short Physical Performance Battery | To be requested | |
| Duke Activity Status Index (DASI) | To be requested | |
| Rapid Eating Assessment for Patients | To be requested | |
| Gestational Age Classification | More information needed | |
| Intrauterine Growth Retardation (IUGR) | To be requested | |
| Gestational Age by Exam (Ballard, Dubowitz version) | To be requested | |
| World Health Organization Quality of Life HIV BREF (WHOQoL-HIV BREF) | To be requested | |
| Center for Epidemiologic Studies Depression Scale (CESD) | To be requested |
8 Analysis Data
This section demonstrates the use of ADaM in HIV in a limited set of use cases:
For information and specifications for the content of datasets that should be submitted as part of an application for drugs intended to treat HIV, please refer to the FDA's HIV Technical Specifications Guidance: https://www.fda.gov/downloads/ForIndustry/DataStandards/StudyDataStandards/UCM603323.pdf .
8.1 Subject Level Analysis Data (ADSL)
The example below shows an abbreviated ADSL dataset, which includes the variables that may be of specific interest to HIV snapshot analysis. Discontinuation status and reason for discontinuation from study have been included for this purpose. Variables representing subject-level data that would commonly occur in ADSL regardless of therapeutic area (e.g., sex, race, age, age groups, geographic region, population flags, treatment start and stop date), are not shown.
Example
The following tables provide examples of an abbreviated ADSL, ADSL dataset metadata, and ADSL variable metadata. The source derivation metadata for the variables are provided for illustrative purposes and not intended to represent standard derivation logic. Additional text in the Source/Derivation/Comment column is meant to provide further discussion for the variable and would not be present in an actual define.xml document.
Example metadata at the dataset and variable level appear below.
ADSL Dataset Metadata
| Dataset | Description | Class | Structure | Purpose | Keys | Location | Documentation |
|---|---|---|---|---|---|---|---|
| ADSL | Subject-Level Analysis Dataset | SUBJECT LEVEL ANALYSIS DATASET | 1 record per subject | Analysis | STUDYID, USUBJID |
adsl.xpt | ADSL.SAS/SAP |
ADSL Variable Metadata
| Variable Name | Variable Label | Type | Codelist/ControlledTerms | Source/Derivation/Comment |
|---|---|---|---|---|
| EOSSTT | End of Study Status | Char | COMPLETED, DISCONTINUED, ONGOING | The subject's status as of the end of study or data cutoff. Examples: COMPLETED, DISCONTINUED, ONGOING. |
| DCSREAS | Reason for Discontinuation from Study | Char | Death, Protocol Deviation, Lack of Efficacy | Reason for subject's discontinuation from study. The source would most likely be the SDTM DS dataset. Null for subjects who completed the study. |
| TRTSDT | Date of First Exposure to Treatment | Num |
Date of first exposure to treatment for a subject in a study. A numeric version of DM.RFXSTDTC. |
This is an example of the ADSL dataset.
adsl.xpt
| Row | STUDYID | USUBJID | EOSSTT | DCSREAS | TRTSDT |
|---|---|---|---|---|---|
| 1 | ABC-123 | ABC-123-001 | DISCONTINUED | Death | 2017-02-01 |
| 2 | ABC-123 | ABC-123-002 | DISCONTINUED | Protocol Deviation | 2017-02-15 |
| 3 | ABC-123 | ABC-123-003 | DISCONTINUED | Lack of Efficacy | 2017-03-02 |
| 4 | ABC-123 | ABC-123-004 | COMPLETED | 2017-03-15 |
8.2 FDA Snapshot Analysis
FDA snapshot is a virologic primary efficacy endpoint analysis that is primarily based on the HIV RNA viral load at a point in time (in this example it is at 48 weeks). Patients are grouped into 3 categories for analysis based on the viral load value as well as other relevant factors (see below); the values displayed on the summary table are "<50 HIV RNA copies/mL", "≥50 copies/mL", and "No Virologic Data". Cut-off points other than 50 copies/mL may also be used.
It is important to note that the category for subjects having ≥50 copies/mL also includes patients who:
- changed any component of background therapy to a different drug class after the first on-treatment visit; or
- changed components of background therapy that were not permitted per protocol, or who changed any background drug in the regimen before Week 48 due to lack of efficacy; or
- discontinued study drug or who were withdrawn from study before Week 48 due to lack of efficacy or loss of efficacy
For patients categorized as "no virologic data", the reason for lack of data should also be recorded.
More information on the snapshot analysis is available in the FDA guidance (https://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm355128.pdf).
Example
This example is based on the 4 possible scenarios shown in the figure. Each scenario is represented as a subject in the ADEFF example dataset.
Figure. FDA Snapshot Analysis Scenarios
The possible values (and decodes) for AVALC are:
- 1=Virologic success (HIV-RNA < 50 copies/mL)
- 2a=HIV-RNA ≥ 50 copies/mL
- 2b=Discontinued because of virologic failure
- 2c=Discontinued because of other reasons and HIV-1 RNA at the time of discontinuation was ≥ 50 copies/mL
- 2d=Optimized Background Therapy (OBT) changed
- 3a=Discontinued because of adverse event or death
- 3b=Discontinued because of other reasons and HIV-1 RNA at the time of discontinuation was < 50 copies/mL
- 3c=Missing data during the window but on study.
These values are further categorized as 1, 2, or 3, with the corresponding definitions: 1=Virologic success, 2=Virologic failure, 3=No virologic data. For more information, see the FDA guidance for submitting clinical trial data sets (https://www.fda.gov/downloads/ForIndustry/DataStandards/StudyDataStandards/UCM603323.pdf).
Example metadata at the dataset and variable level are shown below.
ADEFF Dataset Metadata
| Dataset | Description | Class | Structure | Purpose | Keys | Location | Documentation |
|---|---|---|---|---|---|---|---|
| ADEFF | Snapshot Efficacy Analysis Dataset | BASIC DATA STRUCTURE | 1 record per subject per parameter per analysis visit | Analysis | STUDYID, USUBJID, PARAMCD, AVISIT | adeff.xpt | ADEFF.SAS/SAP |
ADEFF Variable Metadata
| Variable Name | Variable Label | Type | Codelist/Controlled Terms | Notes |
|---|---|---|---|---|
| STUDYID | Study Identifier | Char | ADSL.STUDYID | |
| USUBJID | Unique Subject Identifier | Char | ADSL.USUBJID | |
| PARAM | Parameter | Num | Snapshot Status for cut-point of 50 | The description of the analysis parameter |
| PARAMCD | Parameter Code | Char | SS50 | The short name of the analysis parameter in PARAM |
| AVISIT | Analysis Visit | Char | Week 48 | Analysis Visit based on ADY per windowing defined in the SAP |
| ADT | Analysis Date | Num | The date associated with AVAL and/or AVALC in numeric format. Numeric version of MB.MBDTC | |
| ADY | Analysis Relative Day | Num | The relative day of AVAL and/or AVALC. ADT-TRTSDT+1 | |
| AWTARGET | Analysis Window Target | Num | The target or most desired analysis relative day (ADY) value for a given value of AVISIT | |
| AVALC | Analysis Value (C) | Char | 1;2a;2b;2c;2d;3a;3b;3c | Character analysis value described by PARAM |
| AVALCAT1 | Analysis Value Category 1 | Char | 1;2;3 | A categorization of AVAL or AVALC within a parameter |
| MCRIT1 | Analysis Multi-Response Criterion 1 | Char | Snapshot Table Value | A text string identifying a prespecified criterion within a parameter, where the criterion can have multiple responses |
| MCRIT1ML | Multi-Response Criterion 1 Evaluation | Char | No Virologic Data, HIV-RNA ≥ 50 copies/mL, HIV-RNA < 50 copies/mL | Character variable indicating which level of the criterion defined in MCRITy was met by the data on the record |
| MBSTRESN | Numeric Result/Finding in Standard Units | Num | MB.MBSTRESN | |
| MBSEQ | Sequence Number | Num | MB.MBSEQ | |
| VLIW | Categorization of Viral Load in Window | Char | missing, <50, ≥ 50 | Categorization of MBSTRESN value |
| VFFL | Viral Failure Flag | Char | Y/N | "Y" if VLIW=≥ 50, otherwise "N" |
| DCSREAS | Reason for Discontinuation from Study | Char | ADSL.DCSREAS | |
| DCSFL | Discontinued from Study Flag | Char | Y/N | "Y" if DCSREAS is not null, otherwise "N" Note: Although this is a subject-level value, the inclusion of this variable as a component of the snapshot value depends on the timing of the occurrence. If the occurrence is after the upper limit of the snapshot window, then the value would be set to "N" for this AVISIT value. |
| DCAEDTFL | Discontinued Due to AE or Death Flag | Char | Y/N | "Y" if DCSREAS is in ("Adverse Event", "Death"), otherwise "N" Note: Although this is a subject-level value, the inclusion of this variable as a component of the snapshot value depends on the timing of the occurrence. If the occurrence is after the upper limit of the snapshot window, then the value would be set to "N" for this AVISIT value. |
| DCVFFL | Discontinued Due to Viral Failure Flag | Char | Y/N | "Y" if DCSREAS is in ("Lack of Efficacy"), otherwise "N" Note: Although this is a subject-level value, the inclusion of this variable as a component of the snapshot value depends on the timing of the occurrence. If the occurrence is after the upper limit of the snapshot window, then the value would be set to "N" for this AVISIT value. |
| BKTCHGFL | Background Therapy Change Flag | Char | Y/N | "Y" if background therapy changed after Visit 1, otherwise "N" Note: Although this is a subject-level value, the inclusion of this variable as a component of the snapshot value depends on the timing of the occurrence. If the occurrence is after the upper limit of the snapshot window, then the value would be set to "N" for this AVISIT value. Also note that this value is highly protocol-specific; determining what backgroup therapies may or may not be changed may be defined in each protocol. |
The following is an ADEFF dataset example. Rows 1-4 correspond with Scenarios 1-4 in the preceding figure.
adeff.xpt
| Row | STUDYID | USUBJID | PARAM | PARAMCD | AVISIT | ADT | ADY | AWTARGET | AVALC | AVALCAT1 | MCRIT1 | MCRIT1ML | MBSTRESN | MBSEQ | VLIW | VFFL | DCSREAS | DCSFL | DCAEDTFL | DCVFFL | BKTCHGFL |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ABC-123 | ABC-123-001 | Snapshot Status for cut-point of 50 | SS50 | Week 48 | 336 | 3a | 3 | Snapshot Table Value | No Virologic Data | missing | N | Death | Y | Y | N | N | ||||
| 2 | ABC-123 | ABC-123-002 | Snapshot Status for cut-point of 50 | SS50 | Week 48 | 336 | 2d | 2 | Snapshot Table Value | HIV-RNA ≥ 50 copies/mL | missing | N | Protocol Deviation | Y | N | N | Y | ||||
| 3 | ABC-123 | ABC-123-003 | Snapshot Status for cut-point of 50 | SS50 | Week 48 | 2018-02-04 | 340 | 336 | 2b | 2 | Snapshot Table Value | HIV-RNA ≥ 50 copies/mL | 40 | 46 | <50 | N | Lack of Efficacy | Y | N | Y | N |
| 4 | ABC-123 | ABC-123-004 | Snapshot Status for cut-point of 50 | SS50 | Week 48 | 2018-02-22 | 345 | 336 | 1 | 1 | Snapshot Table Value | HIV-RNA < 50 copies/mL | 40 | 52 | <50 | N | N | N | N | N |
8.3 Pregnancy Outcomes Analysis
This example deals with a scenario in which both mother and infant are considered study subjects. Most of the efficacy outcomes are based on either mother-related topics or infant-related topics. However, 1 of the outcomes is based on comparisons of mother/infant combinations.
The sample datasets are designed to support the pairwise analysis of the mother/infant as a unit. Adverse Pregnancy Outcome is determined based on events that may occur in the mother only or the infant only. These events include spontaneous abortion, fetal death, preterm delivery, or small for gestational age.
One dataset (ADOUTEVT) is designed as per ADaM basic data structure (BDS), with parameters capturing "Y"/"N" for each of the relevant components used in the calculations of the endpoint measures. The second dataset (ADOUTCOM) is designed as an ADAM OTHER, with parameters capturing "Y"/"N" for the endpoint measure. Splitting ADOUTCOM from ADOUTEVT allows for traceability from the ADOUTCOM record back to the source records from the predecessor dataset (ADOUTEVT). Capturing the record in ADOUTEVT only would cause confusion when the outcome record is based on both the mother and infant records because there is no way to determine which USUBJID value should be used in the outcome row.
Known Issue: The ADaM BDS class currently is designed to support subject-level analysis (as well as site-level analysis). However, grouping a number of subjects as an analysis unit has not been addressed. This example proposes the variable USUBJGR1 (USUBJGRy as a generic variable) to allow for the grouping of subjects for analysis purposes. This allows for the calculation of a single analysis value that is based on the data from more than one subject. Because the ADOUTCOM dataset is summarized by family based on the values across multiple subjects, a structure based on family and parameter is needed—which does not fit within the BDS definition. Therefore, it is proposed as an ADAM OTHER class of dataset.
Example metadata at the dataset and variable level for ADOUTEVT are shown below.
ADOUTEVT Dataset Metadata
| Dataset | Description | Class | Structure | Purpose | Keys | Location | Documentation |
|---|---|---|---|---|---|---|---|
| ADOUTEVT | Mother-Child Outcomes Events AD | BASIC DATA STRUCTURE | 1 record per subject per parameter | Analysis | STUDYID, USUBJID, PARAMCD | adoutevt.xpt | ADOUTEVT.SAS/SAP |
ADOUTEVT Variable Metadata
| Variable Name | Variable Label | Type | Controlled Terms / Format | Notes |
|---|---|---|---|---|
| USUBJGR1 | Unique Subject Identifier Group 1 | Char | Assigned: Used to group mothers and infants in the same family. | |
| USUBJID | Unique Subject Identifier | Char | Predecessor: ADSL.USUBJID | |
| PARAM | Parameter | Char | Spontaneous Abortion;Fetal Death;Preterm Delivery;Small for Gestational Age | Assigned: The description of the analysis parameter. |
| PARAMCD | Parameter Code | Char | SPONAB;FETALDTH;PTD;SGESTAGE | Assigned: The short name of the analysis parameter in PARAM. |
| AVALC | Analysis Value (C) | Char | Y;N | Derived: Character analysis value described by PARAM.
|
This is an example of the ADOUEVT dataset.
adoutevt.xpt
| USUBJGR1 | USUBJID | PARAM | PARAMCD | AVALC |
|---|---|---|---|---|
| FAMILY1 | MOTHER1 | Spontaneous Abortion | SPONAB | N |
| FAMILY1 | MOTHER1 | Fetal Death | FETALDTH | N |
| FAMILY1 | INFANT1 | Preterm Delivery | PTD | Y |
| FAMILY1 | INFANT1 | Small for Gestational Age | SGESTAGE | Y |
| FAMILY2 | MOTHER2 | Spontaneous Abortion | SPONAB | N |
| FAMILY2 | MOTHER2 | Fetal Death | FETALDTH | N |
| FAMILY2 | INFANT2 | Preterm Delivery | PTD | N |
| FAMILY2 | INFANT2 | Small for Gestational Age | SGESTAGE | N |
Example metadata at the dataset and variable level for ADOUTCOM are shown below.
ADOUTCOM Dataset Metadata
| Dataset | Description | Class | Structure | Purpose | Keys | Location | Documentation |
|---|---|---|---|---|---|---|---|
| ADOUTCOM | Mother-Child Outcomes Analysis Dataset | ADAM OTHER | 1 record per family per parameter | Analysis | STUDYID, USUBJGR1, PARAMCD | adoutcom.xpt | ADOUTCOM.SAS/SAP |
ADOUTCOM Variable Metadata
| Variable Name | Variable Label | Type | Controlled Terms / Format | Notes |
|---|---|---|---|---|
| USUBJGR1 | Unique Subject Identifier Group 1 | Char | Predecessor: ADOUTEVT.USUBJGR1 | |
| PARAM | Parameter | Char | Adverse Pregnancy Outcome | Assigned: The description of the analysis parameter. |
| PARAMCD | Parameter Code | Char | APOUTCOM | Assigned: The short name of the analysis parameter in PARAM. |
| AVALC | Analysis Value (C) | Char | Y;N | Derived: Character analysis value described by PARAM.
|
This is an example of the ADOUTCOM dataset.
adoutcom.xpt
| USUBJGR1 | PARAM | PARAMCD | AVALC |
|---|---|---|---|
| FAMILY1 | Adverse Pregnancy Outcome | APOUTCOM | Y |
| FAMILY2 | Adverse Pregnancy Outcome | APOUTCOM | N |
Appendices
Appendix A: HIV Standards Team
| Name | Institution/Organization |
|---|---|
| Laura Butte, Team Lead | Critical Path Institute |
| Amy Palmer, Team Lead | CDISC |
| Diane Corey | Critical Path Institute |
| Jane Diefenbach | PharmaStat |
| Kristine Donaty | Statistical Center for HIV/AIDS Research & Prevention (SCHARP) |
| Carrie Fry | Frontier Science and Technology Research Foundation, Inc. |
| Nate Freimark | The Griesser Group |
| Laura Hovind | Frontier Science and Technology Research Foundation, Inc. (FSTRF) |
| Minhee Kang | Center for Biostatistics in AIDS Research (CBAR) (https://www.hsph.harvard.edu/cbar/) |
| Maura Kush | Statistical Center for HIV/AIDS Research & Prevention (SCHARP) |
| Bess LeRoy | CDISC |
| Erin Muhlbradt | NCI-EVS |
| Jon Neville | CDISC |
| Daniel Olson | Critical Path Institute |
| Heather Ribaudo | Center for Biostatistics in AIDS Research (CBAR) (https://www.hsph.harvard.edu/cbar/) |
| Anthony Rodgers | Merck |
| Ed Slezinger | Statistical Center for HIV/AIDS Research & Prevention (SCHARP) |
| Heather Sprenger | Frontier Science and Technology Research Foundation, Inc. (FSTRF) |
| Alana St. Clair | CDISC |
| Sarah Strobino | Frontier Science and Technology Research Foundation, Inc. (FSTRF) |
| Diane Wold | CDISC |
| FDA Team Members | Division |
| Wendy Carter | Division of Antiviral Products |
| Damon Deming | Division of Antiviral Products |
| Wen Zeng | Office of Biometrics, Division IV |
Appendix B: Glossary and Abbreviations
| ADaM | Analysis Data Model |
| ADaMIG | Analysis Data Model Implementation Guide |
| AIDS | Acquired Immunodeficiency Syndrome |
| ART | Antiretroviral Therapy |
| ARV | Antiretroviral |
| BMD | Bone Mineral Density |
| CDASH | Clinical Data Acquisition Standards Harmonization |
| CDC | U.S. Centers for Disease Control and Prevention |
| CDISC | Clinical Data Interchange Standards Consortium |
| CFAST | Coalition for Accelerating Standards and Therapies |
| Collected | Refers to information that is recorded and/or transmitted to the sponsor, including data entered by the site on CRFs/eCRFs as well as vendor data such as core lab data. This term is a synonym for captured. |
| Controlled Terminology | A finite set of values that represent the only allowed values for a data item. These values may be codes, text, or numeric. A codelist is one type of controlled terminology. |
| CRF, aCRF | Case report form (sometimes called a case record form), annotated case report form. A printed, optical, or electronic document designed to record all required information to be reported to the sponsor for each trial subject. |
| cSDRG | Clinical Study Data Reviewers Guide |
| DEXA or DXA | Dual-energy X-Ray Absorptiometry |
| Domain | A collection of observations with a topic-specific commonality about a subject |
| Embryo | Early stage in the prenatal development of an animal. This stage occurs from implantation until closure of the hard palate. |
| Fetus | Late stage in the prenatal development of an animal. This stage occurs from the closure of the hard palate until birth. |
| HIV | Human Immunodeficiency Virus |
| IAS–USA | International Antiviral Association–USA |
| Infant | A young child between 1 month and 2 years of age. |
| NCI EVS | National Cancer Institute Enterprise Vocabulary Services |
| Newborn | An infant during the first month after birth. |
| OBT | Optimized Background Therapy |
| PrEP | Pre-exposure Prophylaxis |
| SDTM | Study Data Tabulation Model |
| SDTMIG | SDTM Implementation Guide (for Human Clinical Trials) |
| STI | Sexually Transmitted Infection |
| Subject | A participant in a study. |
| VISR | Vaccine-induced Seroreactivity |
Appendix C: Non-Standard Variables
The following table lists the non-standard variables used in this document, and gives their parent domain and variable-level metadata.
(Parenthesis indicates CDISC/NCI codelist)
Appendix D: References
Works Cited
- HIV/AIDS online Q&A. World Health Organization. http://www.who.int/features/qa/71/en. Updated November 2017. Accessed March 27, 2018.
- Terms, definitions, and calculations used in CDC HIV surveillance publications. Centers for Disease Control and Prevention. Updated December 12, 2016. Accessed November 29, 2018.
- World Health Organization. Guideline on when to start antiretroviral therapy and on pre-exposure prophylaxis for HIV. http://apps.who.int/iris/bitstream/10665/186275/1/9789241509565_eng.pdf. Published September 2015. Accessed March 27, 2018.
- HIV treatment, the viral reservoir, and HIV DNA. National Institute of Allergy and Infectious Diseases. https://www.niaid.nih.gov/diseases-conditions/hiv-treatment-viral-reservoir-hiv-dna. Updated March 6, 2018. Accessed March 27, 2018.
- HIV/AIDS: the basics. U.S. Department of Health and Human Services. https://aidsinfo.nih.gov/understanding-hiv-aids/fact-sheets/19/45/hiv-aids--the-basics. Updated November 6, 2018. Accessed November 27, 2018.
- About HIV/AIDS. Centers for Disease Control and Prevention. https://www.cdc.gov/hiv/basics/whatishiv.html. Updated March 16, 2018. Accessed March 27, 2018.
- HIV/AIDS fact sheet. World Health Organization. http://www.who.int/mediacentre/factsheets/fs360/en/. Updated November 2017. Accessed March 27, 2018.
- LGBTQ+ definitions. Trans Student Educational Resources. http://www.transstudent.org/definitions. Published 2018. Accessed March 27, 2018.
- Understanding gender. Gender Spectrum. https://www.genderspectrum.org/quick-links/understanding-gender/. Published 2017. Accessed March 27, 2018.
- Menstrual changes. The Well Project. http://www.thewellproject.org/hiv-information/menstrual-changes. Published September 12, 2017. Accessed March 28, 2018.
- US Department of Health and Human Services, Food and Drug Administration. Human Immunodeficiency Virus-1 Infection: Developing Antiretroviral Drugs for Treatment. Guidance for Industry. Washington, DC: US Department of Health and Human Services; 2015. https://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm355128.pdf. Accessed November 27, 2018.
- Drug resistance mutations in HIV-1. International Antiviral Society–USA. https://www.iasusa.org/content/drug-resistance-mutations-in-HIV. Accessed March 27, 2018.
- McComsey GA, Tebas P, Shane E, et al. Bone disease in HIV infection: a practical review and recommendations for HIV care providers. Clin Infect Dis. 2010;51(8):937-946. doi:10.1086/656412.
Additional Reading
-
Grady D. Vaginal ring with drug lowers H.I.V. rates in African women. New York Times. February 22, 2016. https://www.nytimes.com/2016/02/23/health/vaginal-ring-hiv-aids-drug-dapivirine.html. Accessed March 27, 2018.
Appendix E: Representations and Warranties, Limitations of Liability, and Disclaimers
CDISC Patent Disclaimers
It is possible that implementation of and compliance with this standard may require use of subject matter covered by patent rights. By publication of this standard, no position is taken with respect to the existence or validity of any claim or of any patent rights in connection therewith. CDISC, including the CDISC Board of Directors, shall not be responsible for identifying patent claims for which a license may be required in order to implement this standard or for conducting inquiries into the legal validity or scope of those patents or patent claims that are brought to its attention.
Representations and Warranties
"CDISC grants open public use of this User Guide (or Final Standards) under CDISC's copyright."
Each Participant in the development of this standard shall be deemed to represent, warrant, and covenant, at the time of a Contribution by such Participant (or by its Representative), that to the best of its knowledge and ability: (a) it holds or has the right to grant all relevant licenses to any of its Contributions in all jurisdictions or territories in which it holds relevant intellectual property rights; (b) there are no limits to the Participant's ability to make the grants, acknowledgments, and agreements herein; and (c) the Contribution does not subject any Contribution, Draft Standard, Final Standard, or implementations thereof, in whole or in part, to licensing obligations with additional restrictions or requirements inconsistent with those set forth in this Policy, or that would require any such Contribution, Final Standard, or implementation, in whole or in part, to be either: (i) disclosed or distributed in source code form; (ii) licensed for the purpose of making derivative works (other than as set forth in Section 4.2 of the CDISC Intellectual Property Policy ("the Policy")); or (iii) distributed at no charge, except as set forth in Sections 3, 5.1, and 4.2 of the Policy. If a Participant has knowledge that a Contribution made by any Participant or any other party may subject any Contribution, Draft Standard, Final Standard, or implementation, in whole or in part, to one or more of the licensing obligations listed in Section 9.3, such Participant shall give prompt notice of the same to the CDISC President who shall promptly notify all Participants.
No Other Warranties/Disclaimers. ALL PARTICIPANTS ACKNOWLEDGE THAT, EXCEPT AS PROVIDED UNDER SECTION 9.3 OF THE CDISC INTELLECTUAL PROPERTY POLICY, ALL DRAFT STANDARDS AND FINAL STANDARDS, AND ALL CONTRIBUTIONS TO FINAL STANDARDS AND DRAFT STANDARDS, ARE PROVIDED "AS IS" WITH NO WARRANTIES WHATSOEVER, WHETHER EXPRESS, IMPLIED, STATUTORY, OR OTHERWISE, AND THE PARTICIPANTS, REPRESENTATIVES, THE CDISC PRESIDENT, THE CDISC BOARD OF DIRECTORS, AND CDISC EXPRESSLY DISCLAIM ANY WARRANTY OF MERCHANTABILITY, NONINFRINGEMENT, FITNESS FOR ANY PARTICULAR OR INTENDED PURPOSE, OR ANY OTHER WARRANTY OTHERWISE ARISING OUT OF ANY PROPOSAL, FINAL STANDARDS OR DRAFT STANDARDS, OR CONTRIBUTION.
Limitation of Liability
IN NO EVENT WILL CDISC OR ANY OF ITS CONSTITUENT PARTS (INCLUDING, BUT NOT LIMITED TO, THE CDISC BOARD OF DIRECTORS, THE CDISC PRESIDENT, CDISC STAFF, AND CDISC MEMBERS) BE LIABLE TO ANY OTHER PERSON OR ENTITY FOR ANY LOSS OF PROFITS, LOSS OF USE, DIRECT, INDIRECT, INCIDENTAL, CONSEQUENTIAL, OR SPECIAL DAMAGES, WHETHER UNDER CONTRACT, TORT, WARRANTY, OR OTHERWISE, ARISING IN ANY WAY OUT OF THIS POLICY OR ANY RELATED AGREEMENT, WHETHER OR NOT SUCH PARTY HAD ADVANCE NOTICE OF THE POSSIBILITY OF SUCH DAMAGES.
Note: The CDISC Intellectual Property Policy can be found at: cdisc_policy_003_intellectual_property_v201408.pdf.