In most cases, viewing the annotated Case Report Form (aCRF) example with metadata is straightforward when looking at the same data transformed into SDTM. In some instances, however, it is not. In these cases, "magic" happens from data collection in the CDASH example to the data tabulation in the SDTM example. There are assumptions made regarding the collected data and metadata that are then transformed to the submission data tables. It is not necessarily clear how one went from point A to point C. This article provides a look behind the curtain at the magic that sometimes occurs between the CDASH and SDTM examples.
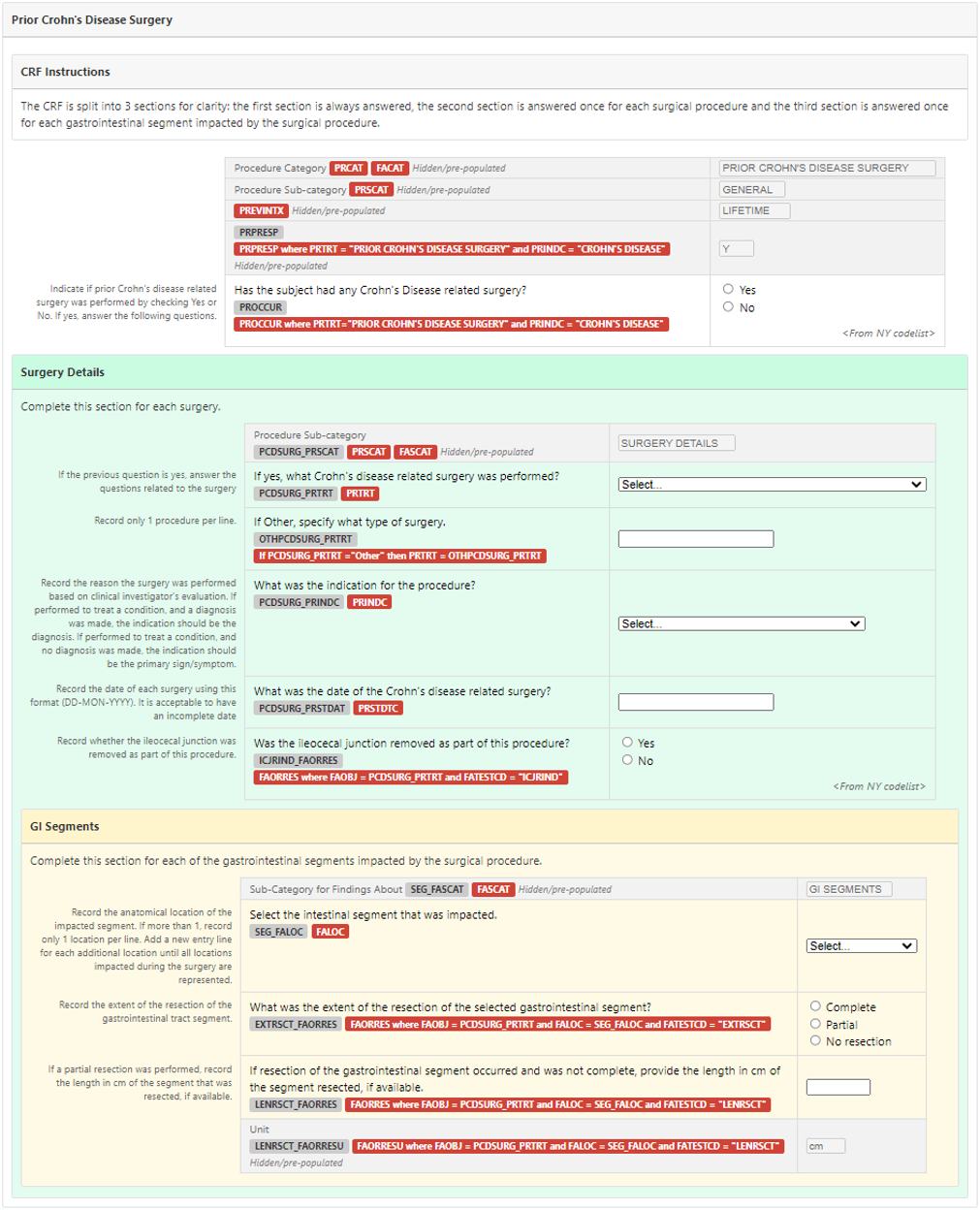
Prior Crohn's Disease Surgery
CRF Instructions
The CRF is split into three sections for clarity: the first section is always answered, the second is answered once for each surgical procedure and the third is answered once for each gastrointestinal segment impacted by the surgical procedure.
Example 1
In this example study, the sponsor was interested in collecting information on any Crohn's disease-related surgeries.
The drop-down list is not visible in the screenshot below but can be seen in the CRF metadata table included in the Crohn’s Therapeutic Area User Guide. Note: the drop-down list is not exhaustive, but rather details the more common procedures associated with Crohn's disease. Depending on the type of procedure, the remaining questions are displayed when applicable.
CDASH Data Representations The following CDASH data representations have been included as example structures to help visualize data that might be collected using the example CRF shown above. These data representations are included for illustration purposes only and should not be taken as recommendations for CDASH data structures or EDC system design. In each of the data representation tables shown below:
- "Context" or "header" information that is not explicitly collected in the example CRF, but would be assigned in the EDC system, has been included in each of the data representation tables in greyed out columns. This includes the study identifier (STUDYID), the subject identifier (SUBJID), the visit name (VISIT) and the visit date (VISDAT).
- Apart from the context/header columns—and some additional key columns for the first example structure (which are described in more detail below)—only the CDASH variables specified in the CRF metadata are shown. SDTM variables referenced in SDTM Target Variable Mapping annotations are not included in these CDASH data representations.
- Data values are represented as collected or defined in the CRF metadata:
- Pre-populated values are included on every record in the format specified in the CRF metadata.
- Permissible values are represented in the format specified in the metadata:
- If the CRF metadata includes the controlled terminology value (or "database value") associated with each displayed permissible value, the specified controlled terminology value is used in the data representation table. For example, when the permissible values are "Y=Yes; N=No", the data representation table will contain "Y" or "N".
- If the CRF metadata only includes displayed permissible values, these values are included, as specified, in the data representation table. For example, when the permissible values are "Complete; Partial; No resection", the data representation table will contain "Complete", "Partial" or "No resection".
- Date values are represented in the collection format specified in the CRF completion instructions (DD-MON-YYYY).
- Free text was collected in upper case.
- The coloring of the cells corresponds with the coloring of the three sections in the example CRF shown above:
- White cells contain information from the non-repeating "Prior Crohn's Disease Surgery" CRF section.
- Green cells contain information from the repeating "Surgery Details" CRF section.
- Yellow cells contain information from the repeating "GI Segments" CRF section.
- The example collected data values correspond with the values represented in the SDTM example datasets.
Vertical Structure - Separate Tables
Data from each section of the CDASH CRF might be recorded in a separate database table.
To retain the relationship between the information recorded in each of the sections, additional key columns might be added in the EDC system. In this example, the tables created by the EDC system include a CRFSECTION_NAME column to identify the name of the CRF section used to collect the data and a CRFSECTION_ID column, which was populated with an incrementing number for each repeat of the section. For non-repeating sections, CRFSECTION_ID was populated with 0. To describe nesting of CRF sections, the EDC system tables also include PARENT_CRFSECTION_NAME and PARENT_CRFSECTION_ID columns to identify the name and, when relevant, repeat number of the parent CRF section. For top-level CRF sections, which have no parent, these columns are blank.
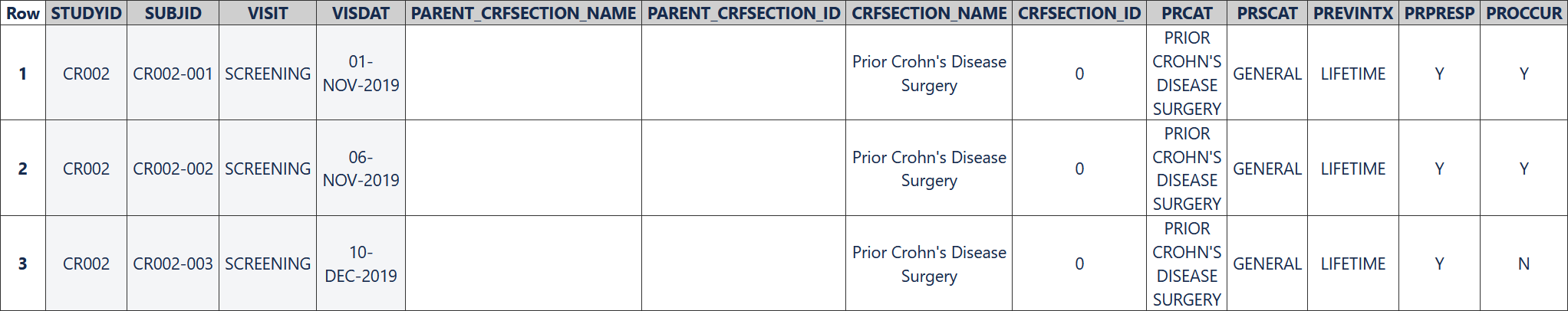
The following table represents data collected for three subjects using the first, non-repeating "Prior Crohn's Disease Surgery" CRF section.
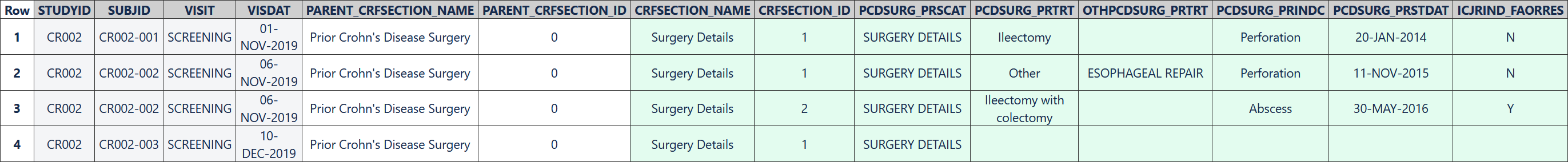
The following table represents data collected for three subjects using the repeating "Surgery Details" CRF section. There was one surgery collected for subject CR002-001 and two collected for subject CR002-002. No surgery was collected for subject CR002-003, but the example EDC system generated a blank row in the table.
Row 1: Shows the collected details of the first surgery recorded for subject CR002-001.
Row 2: Shows the collected details of the first surgery recorded for subject CR002-002.
Row 3: Shows the collected details of the second surgery recorded for subject CR002-002.
Row 4: Was created automatically by the EDC system. All enterable variables (PCDSURG_PRTRT, OTHPCDSURG_PRTRT, PCDSURG_PRINDC, PCDSURG_PRSTDAT and ICJRIND_FAORRES) are blank because the subject CR002-003 did not have any prior surgery for Crohn's disease.
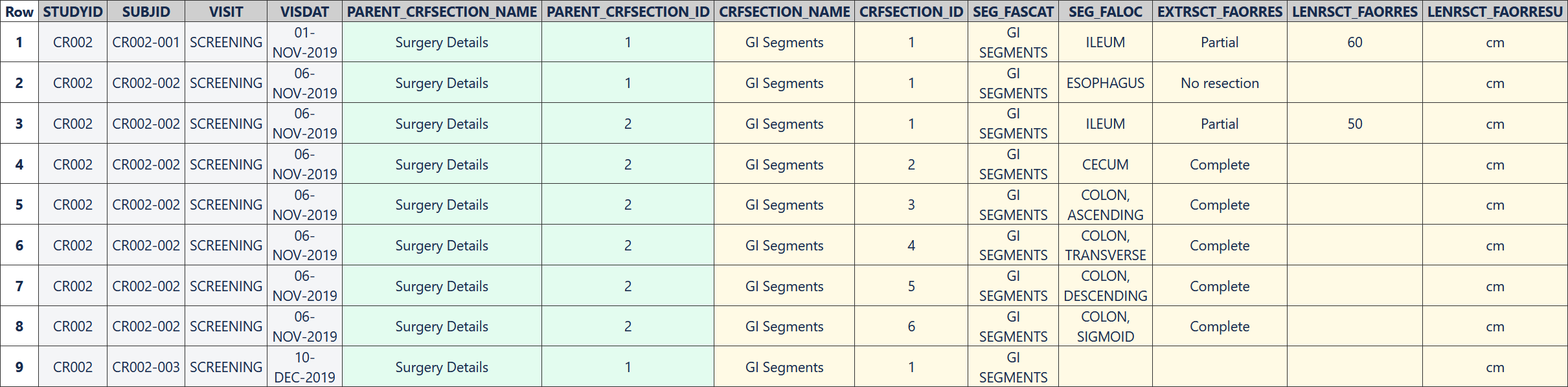
The following table represents data collected for three subjects using the repeating "GI Segments" CRF section. Information about only one impacted gastrointestinal segment was collected for both the single surgery entered for subject CR002-001 and the first surgery entered for subject CR002-002, and information about six GI segments was collected for the second surgery entered for subject CR002-002. No GI segment details were collected for subject CR002-003, but the example EDC system generated a blank row in the table.
Row 1: Shows the collected details of the single gastrointestinal segment that was impacted by the first surgery recorded for subject CR002-001.
Row 2: Shows the collected details of the single gastrointestinal segment that was impacted by the first surgery recorded for subject CR002-002.
Rows 3-8: Show the collected details of the six gastrointestinal segments that were impacted by the second surgery recorded for subject CR002-002.
Row 9: Was created automatically by the EDC system. The enterable variables (SEG_FALOC, EXTRSCT_FAORRES and LENRSCT_FAORRES) are blank, but values are present for the variables that were pre-populated on the CRF (SEG_FASCAT and LENRSCT_FAORRESU).
Vertical Structure - Single Table
Alternatively, data collected in all three sections of the CRF might be stored in a single database table, with higher-level information repeated on each of the rows containing information from a lower level. The following table contains the same example data that is represented in the separate tables above.
Row 1: Shows the collected details of the single gastrointestinal segment that was impacted by the first surgery recorded for subject CR002-001.
Row 2: Shows the collected details of the single gastrointestinal segment that was impacted by the first surgery recorded for subject CR002-002.
Rows 3-8: Show the collected details of the six gastrointestinal segments that were impacted by the second surgery recorded for subject CR002-002. The information entered on the non-repeating CRF section and the details of the "Ileectomy with colectomy" parent procedure (which were entered on the second repeat of the repeating "Surgery Details" CRF section) are repeated on the records for all gastrointestinal segments impacted by the "Ileectomy with colectomy" procedure.
Row 9: Shows the information collected in the non-repeating "Prior Crohn's Disease Surgery" CRF section. As there was no prior surgery for Crohn's disease for this subject, no further information was collected on the repeating CRF sections. For these sections, the enterable variables (PCDSURG_PRTRT, OTHPCDSURG_PRTRT, PCDSURG_PRINDC, PCDSURG_PRSTDAT, SEG_FALOC, EXTRSCT_FAORRES, LENRSCT_FAORRES and ICJRIND_FAORRES) are blank, but values are present for the variables that were pre-populated on the CRF (PCDSURG_PRSCAT, SEG_FASCAT and LENRSCT_FAORRESU).
In some EDC systems, collected data might be stored in a horizontal structure, with all data collected on the CRF for single subject being represented on a single record. In such a structure, the set of values collected for each completion of the repeating CRF sections would be represented in a separate set of variables (e.g., PCDSURG1_PRTRT, PCDSURG1_PRSTDAT for details of the first surgery and PCDSURG2_SEG_FALOC3, PCDSURG2_LENRSCT_FAORRES3, etc. for details of the third GI segment impacted by the second surgery). A representation of this data structure is not included here because the variable names would not align with the CDASH variable names defined in the example CRF metadata above.
Mapping to SDTM
When data from the CDASH data structures represented above are mapped into SDTM datasets, some restrictions and conversions would need to be applied to create SDTM conformant data, including:
- Mapping subsets of the collected data to appropriate domains.
- Exclusion of missing data: SDTM records would generally not be created if a data value has not been collected.
- Addition of supporting information specified in SDTM Target Variable Mapping annotations. If the SDTM Target Variable Mapping annotation references a value for an SDTM variable that does not originate on the CRF (e.g., where PRTRT = "PRIOR CROHN'S DISEASE SURGERY" and PRINDC = "CROHN'S DISEASE" for the PRPRESP and PROCCUR CDASH variables), this indicates that the specified SDTM variable(s) should be included in the SDTM target dataset and populated with the specified value(s) on the record that contains the value mapped from the CDASH variable.
- Conversion of some text values to upper case.
- Conversion of date values to ISO 8601 format.
- When necessary, mapping of values generated by the EDC system into appropriate SDTM values. For example, values from CRFSECTION_ID could be used to populate PRLNKID and values from PARENT_CRFSECTION_ID could be used to populate FALNKID when creating linked PR and FAPR records from the "Surgery Details" and "GI Segments" CRF sections respectively.
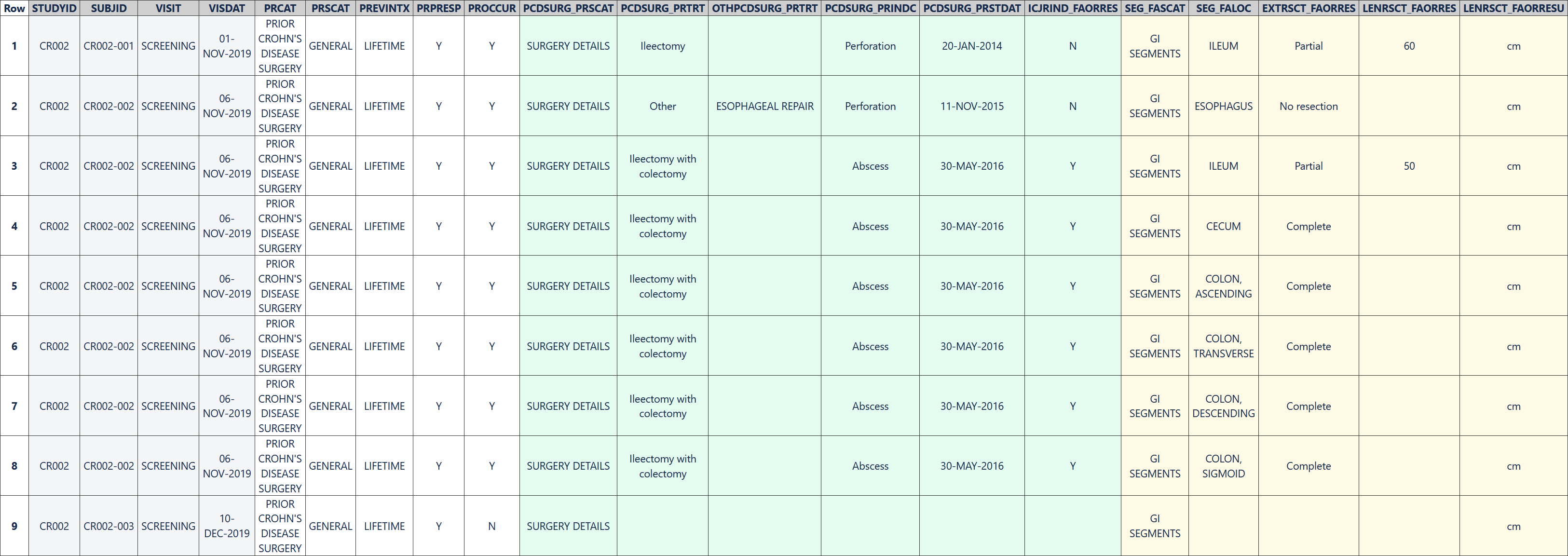
In this example, the sponsor was interested in prior surgery related to Crohn's disease. The sponsor elected to include in the PR domain a record indicating whether or not each subject had had any prior surgery related to Crohn's disease. On these records, PRPRESP is "Y" to indicate that the type of surgery ("prior Crohn's disease surgery") was pre-specified on the CRF and PROCCUR indicates whether or not the type of surgery had occurred for the subject. Other sponsors may elect not to include such a record and instead use the PRYN CDASH variable for data cleaning without submitting the response in the SDTM dataset.
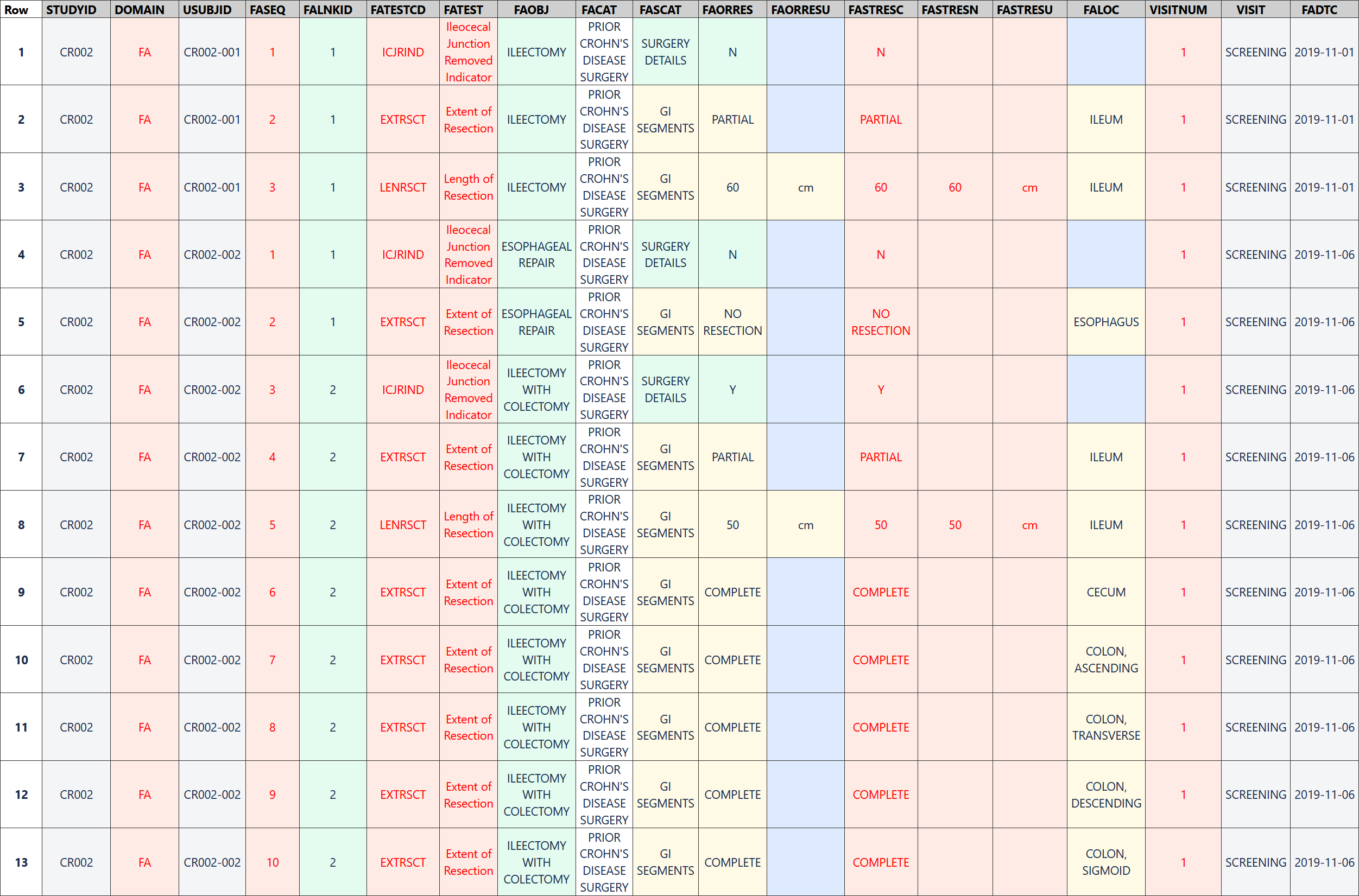
When prior Crohn's disease surgery had occurred, the sponsor also collected details of each surgery that was performed. For most surgeries, the name of the surgery was selected from a pre-specified list on the CRF to ensure the same terminology was used and to avoid data entry errors. When the required surgery name was not present in the drop-down list, "Other" was chosen from the list and the name of the surgery was entered as free text. Although the terms in the drop-down list are well-defined, the investigator was still asked to provide details of any resection for each GI segment impacted by the surgery to verify the presence or absence of each segment. These details were recorded in the FAPR dataset where the FATESTCD and FATEST variables indicate the information collected and the FALOC variable indicates the GI segment.
In the dataset below, certain Expected variables have been omitted in consideration of space and clarity.
The SDTM data representations have been color coded to help visualize data collected using the example CRF shown in the CDASH example and how it would flow into the SDTM dataset.
- "Context" or "header" information that is not explicitly collected in the example CRF, but would be obtained from information assigned in the EDC system, has been included in each of the data representation tables in greyed out columns. This includes the study identifier (STUDYID), the subject identifier (USUBJID), the visit name (VISIT) and the date of data collection (PRDTC), which was obtained from visit date.
- The coloring of the cells indicates the source of any information in the cell:
- As with the CDASH data representation tables above, for cells containing information derived directly from collected data, the coloring corresponds with the coloring of the three sections in the example CRF shown above:
- White cells contain information from the non-repeating "Prior Crohn's Disease Surgery" CRF section.
- Green cells contain information from the repeating "Surgery Details" CRF section. The linking value is colored green to show where the link comes from in the previous data representations.
- Yellow cells contain information from the repeating "GI Segments" CRF section.
- As with the CDASH data representation tables above, for cells containing information derived directly from collected data, the coloring corresponds with the coloring of the three sections in the example CRF shown above:
- Additional coloring:
- Light red cells with red font contain information generated during the SDTM mapping. The empty red cells did not have data to move forward, - for example, when mapping a non-numeric original result to FASTRESN.
- Blue cells are empty because the corresponding source record contained no information to be mapped to the SDTM variable.
- The example SDTM data values correspond with the values represented in the CDASH example datasets above.
Rows 1, 3: Show the subject had prior Crohn's disease surgery.
Row 2: Shows that subject CR002-001 had an ileectomy. The procedure was due to a perforation.
Row 4: Shows that subject CR002-002 had esophageal repair due to perforation.
Row 5: Shows that subject CR002-002 had ileectomy with colectomy due to abscess.
Row 6: Shows that subject CR002-003 had not had any Crohn's disease surgeries in their lifetime.
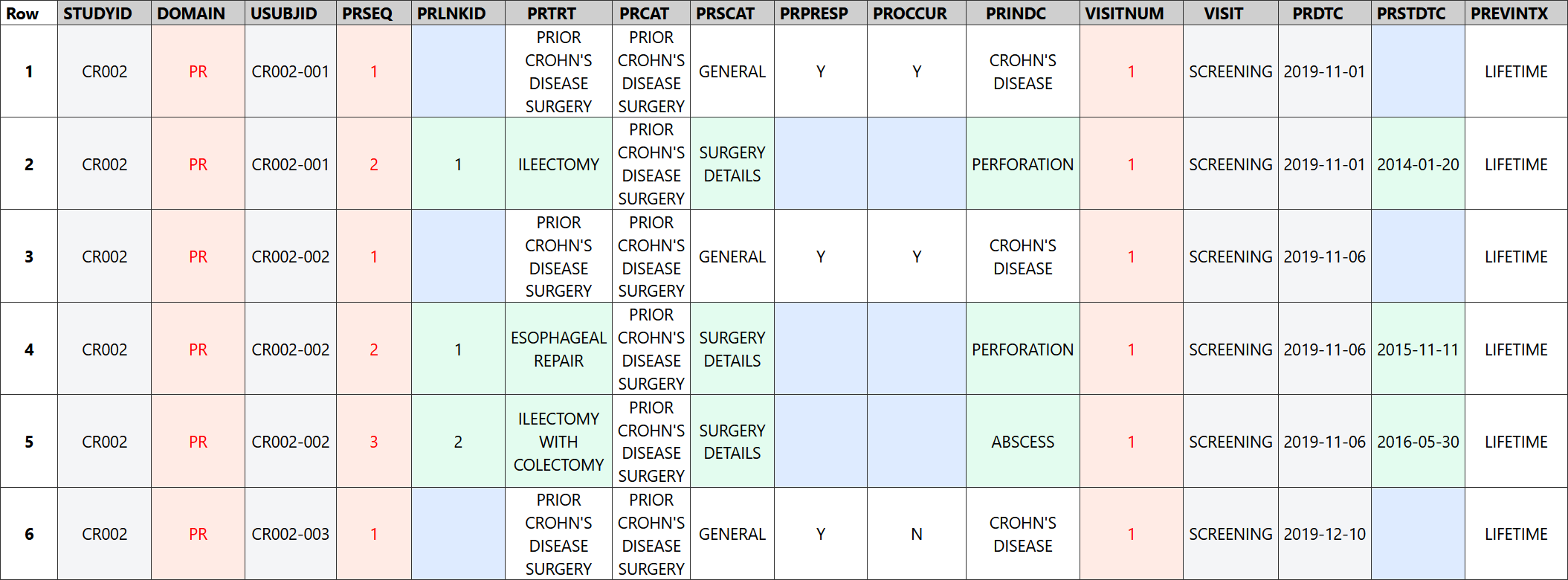
The FA dataset represents details of each of the surgeries that were performed, including the extent of resection of each gastrointestinal segment impacted by the surgery and whether the ileocecal junction was removed during the surgery.
Rows 1-3: Show that the ileocecal junction was not removed during the ileectomy procedure, the subject had a partial resection of the ileum, and the length of the segment resected was 60 cm.
Rows 4-5: Show that the ileocecal junction was not removed during the esophageal repair procedure and there was no resection of the esophagus.
Row 6: Shows the ileocecal junction was removed as part of the ileectomy with colectomy procedure.
Rows 7-13: Show the extent of resection of each segment impacted by the ileectomy with colectomy procedure. Only part of the ileum (50 cm) was removed during this surgical procedure, but the cecum and all parts of the colon were completely removed.
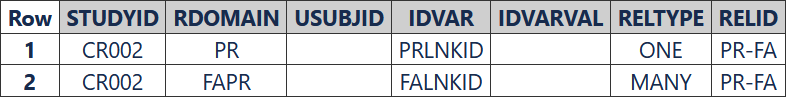
The RELREC dataset shows the relationship between the PR and FAPR datasets. The --LNKID variables were used to relate the records in PR and FAPR.
Depending on how the system is set up and what tool is being used, this may look different. There are several possible ways in which the "magic" could be implemented and, if the transformations are implemented within the system, collected data may never actually be present in separate datasets such as the CDASH data representation tables shown above.
The intention of this article has been to explain how the CDASH variable annotations included in the example CRF align with the potential structure of the collected data, while the SDTM mapping annotations help to indicate how the data gets transformed.
This article aims to help the user understand the transformation of a complex EDC extract. There are variations in the way in which any given EDC system might handle repeating fields or CRF sections. There is also variability among EDC tools; therefore, it is not possible to include all the different representations.
For an overview of the intricacies of the magic that happened between CDASH and SDTM, we invite you to watch our Public Review webinar for Work Package 1 of the Crohn’s Disease Therapeutic Area User Guide, which addresses Additional CDASH to SDTM Data Representation..