The earliest version of the SDTMIG had only one domain for tests on biologic specimens taken from a study subject, the Laboratory Test (LB) domain. SDTMIG v3.4 has 10 domains for specimen-based findings, plus the Biospecimen Events domain. Two of the new domains, Genomics Findings (GF) and Cell Phenotype Findings (CP), as well as the revised Immunogenicity Specimen Findings (IS) domain include domain-specific variables whose meanings are likely not obvious to those who are not subject matter experts in these specialty labs. This decision tree can help an implementer decide where to represent lab data, and a reviewer to locate data.
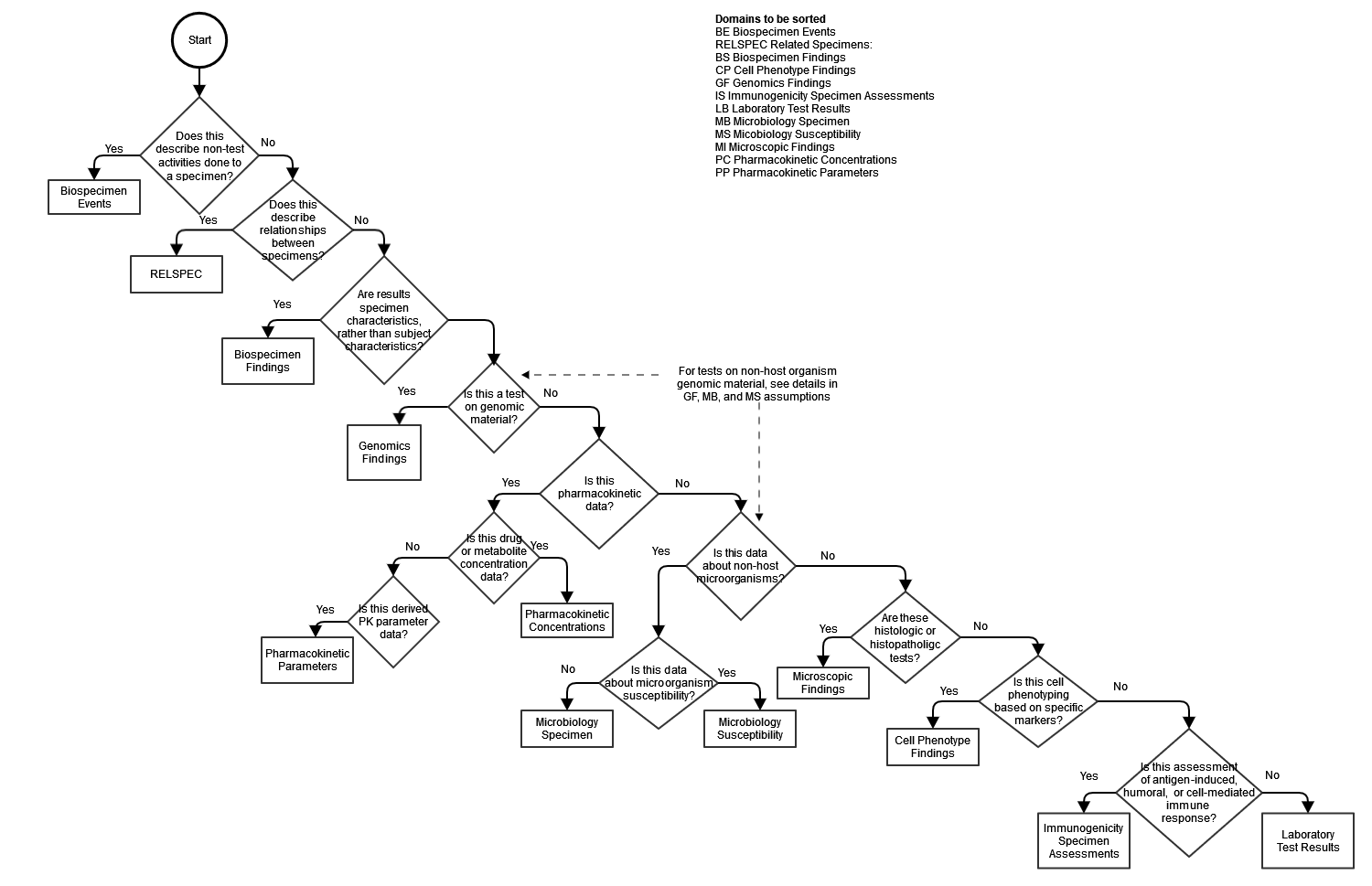
As noted in the diagram, there are some subtleties in deciding where to represent tests on genomic material from non-host organisms, which are explained in detail in the SDTMIG v3.4 in the assumptions for the GF, MB, and MS domains.
From the assumptions for the GF domain:
- Regarding genetic testing on non-host organisms (including, but not limited to bacteria, viruses, and parasites), the following additional assumptions apply:
- Tests that give genetic results (e.g., expressed in terms of genetic variation, specific sequence information) on non-host organisms that have been identified in subject samples should be represented in GF. To distinguish these findings from subject genetic data, the variable NHOID must be populated to identify the non-host organism as the focus of the record (see Section 9.2, Non-host Organism Identifiers , assumption 2 for more information).
- If the purpose of the test is to detect or determine the identity of a viable, non-host organism or infectious agent in a subject sample, data should be represented in the Microbiology Specimen (MB) domain.
- Tests that are used to determine the resistance/susceptibility of a non-host organism to a drug on a genetic basis should be represented in the Microbiology Susceptibility (MS) domain.
- If the test provides both genetic data and susceptibility/resistance data, genetic data should be represented in GF and the susceptibility/resistance data should be represented in the MS domain (See Section 6.3.5.7.2, Microbiology Susceptibility , assumption 1b for more information).
From the assumptions for the MS domain:
- Genetic tests that provide results in terms of susceptible/resistant only (e.g., nucleic acid amplification tests (NAAT)). Genotypic tests that provide results in terms of specific changes to nucleotides, codons, or amino acids of genes/gene products associated with resistance should be represented in the Genomic Findings (GF) domain, as that domain structure contains the variables necessary to accommodate data of this type. If a test provides both mutation data and susceptibility data, the mutation results should be represented in GF and the susceptibility information should be represented in MS. In these cases, the GF records should be linked via RELREC to susceptibility records in MS.
- As in 1.a.ii, MSAGENT should be populated with the drug whose action would be affected by the genetic marker being assessed via the genotypic test. MSCONC and MSCONCU are null in these records.